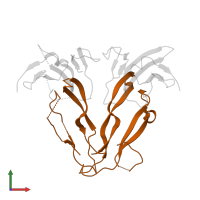Assemblies
Assembly Name:
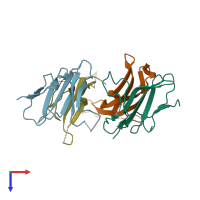
Activin receptor type-2B and Inhibin beta A chain
Multimeric state:
hetero tetramer
Accessible surface area:
19240.03 Å2
Buried surface area:
3000.03 Å2
Dissociation area:
1,500.01
Å2
Dissociation energy (ΔGdiss):
-8.39
kcal/mol
Dissociation entropy (TΔSdiss):
32.26
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-140141
Macromolecules
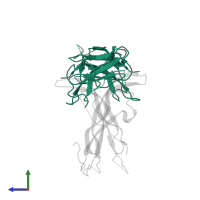
Chains: A, C
Length: 105 amino acids
Theoretical weight: 12.34 KDa
Source organism: Rattus norvegicus
Expression system: Spodoptera frugiperda
UniProt:
Pfam: Activin types I and II receptor domain
InterPro:
SCOP: Extracellular domain of cell surface receptors
Length: 105 amino acids
Theoretical weight: 12.34 KDa
Source organism: Rattus norvegicus
Expression system: Spodoptera frugiperda
UniProt:
- Canonical:
 P38445 (Residues: 19-119; Coverage: 20%)
P38445 (Residues: 19-119; Coverage: 20%)
Pfam: Activin types I and II receptor domain
InterPro:
- Snake toxin-like superfamily
- Ser/Thr protein kinase, TGFB receptor
- Activin types I and II receptor domain
SCOP: Extracellular domain of cell surface receptors
Chains: B, D
Length: 116 amino acids
Theoretical weight: 12.99 KDa
Source organism: Homo sapiens
Expression system: Cricetulus griseus
UniProt:
Pfam: Transforming growth factor beta like domain
InterPro:
SCOP: Transforming growth factor (TGF)-beta
Length: 116 amino acids
Theoretical weight: 12.99 KDa
Source organism: Homo sapiens
Expression system: Cricetulus griseus
UniProt:
- Canonical:
 P08476 (Residues: 311-426; Coverage: 29%)
P08476 (Residues: 311-426; Coverage: 29%)
Pfam: Transforming growth factor beta like domain
InterPro:
- Transforming growth factor-beta, C-terminal
- Cystine-knot cytokine
- Transforming growth factor-beta-related
- Transforming growth factor beta, conserved site
SCOP: Transforming growth factor (TGF)-beta