EC 2.7.7.101: DNA primase DnaG
Reaction catalysed:
SsDNA + n NTP = ssDNA/pppN(pN)(n-1) hybrid + (n-1) diphosphate
Systematic name:
Nucleotide 5'-triphosphate:single-stranded DNA nucleotidyltransferase (DNA-RNA hybrid synthesizing)
Alternative Name(s):
- DnaG
GO terms
Biochemical function:
Biological process:
Cellular component:
- not assigned
Sequence families
Structure domains
 CATH domains
CATH domains
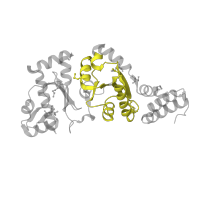
1.20.50.20
Class:
Mainly Alpha
Architecture:
Up-down Bundle
Topology:
Pheromone ER-1
Homology:
DnaG, RNA polymerase domain, helical bundle
Occurring in:

3.40.1360.10
Class:
Alpha Beta
Architecture:
3-Layer(aba) Sandwich
Topology:
Dna Topoisomerase Vi A Subunit; Chain: A, domain 2
Homology:
Dna Topoisomerase Vi A Subunit; Chain: A, domain 2
Occurring in: