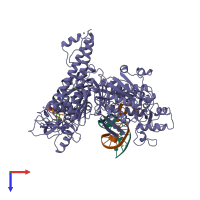
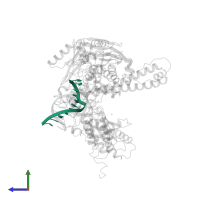
Assemblies
Assembly Name:
DNA-directed DNA polymerase and DNA
Multimeric state:
hetero trimer
Accessible surface area:
45515.46 Å2
Buried surface area:
5086.74 Å2
Dissociation area:
910.6
Å2
Dissociation energy (ΔGdiss):
13.64
kcal/mol
Dissociation entropy (TΔSdiss):
10.11
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-113547
Macromolecules
Chain: A
Length: 903 amino acids
Theoretical weight: 104.74 KDa
Source organism: Escherichia phage RB69
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
Length: 903 amino acids
Theoretical weight: 104.74 KDa
Source organism: Escherichia phage RB69
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q38087 (Residues: 1-903; Coverage: 100%)
Q38087 (Residues: 1-903; Coverage: 100%)
Pfam:
InterPro:
- DNA-directed DNA polymerase T4 type
- Ribonuclease H-like superfamily
- DNA-directed DNA polymerase, family B, exonuclease domain
- DNA-directed DNA polymerase, family B
- Ribonuclease H superfamily
- DNA-directed DNA polymerase, family B, multifunctional domain
- DNA/RNA polymerase superfamily
- DNA polymerase, palm domain superfamily
- DNA-directed DNA polymerase, family B, conserved site
- DNA Polymerase, chain B, domain 1
- Ribonuclease H-like superfamily/Ribonuclease H
- Palm domain of DNA polymerase
- Helix hairpin bin
- Monooxygenase
- DnaQ-like 3'-5' exonuclease
Name:
DNA (5'-D(*GP*CP*GP*GP*AP*AP*CP*TP*AP*CP*T)-3')
Representative chains: E
Source organism: Escherichia phage RB69 [12353]
Expression system: Not provided
Length: 11 nucleotides
Theoretical weight: 3.36 KDa
Representative chains: E
Source organism: Escherichia phage RB69 [12353]
Expression system: Not provided
Length: 11 nucleotides
Theoretical weight: 3.36 KDa
Name:
DNA (5'-D(*AP*GP*TP*AP*GP*TP*TP*CP*CP*GP*CP*G)-3')
Representative chains: D
Source organism: Escherichia phage RB69 [12353]
Expression system: Not provided
Length: 12 nucleotides
Theoretical weight: 3.68 KDa
Representative chains: D
Source organism: Escherichia phage RB69 [12353]
Expression system: Not provided
Length: 12 nucleotides
Theoretical weight: 3.68 KDa