EMD-8481
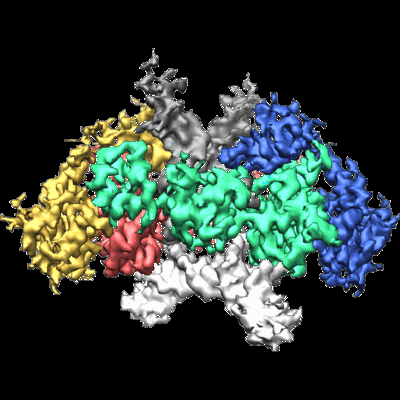
Structure of tetrameric HIV-1 Strand Transfer Complex Intasome
EMD-8481
Single-particle3.9 Å
 Deposition: 28/11/2016
Deposition: 28/11/2016Map released: 11/01/2017
Last modified: 13/03/2024
Concentration: 0.5
mg/mL
Buffer
Grid
Vitrification
Cryogen name: ETHANE
Chamber humidity: 50%
Chamber temperature: 277 K
Instrument: HOMEMADE PLUNGER
Details: Sample containing HIV STC intasomes in SEC buffer was applied onto freshly plasma-treated (6 seconds, Gatan Solarus plasma cleaner) holey gold UltrAuFoil grids (Quantifoil), adsorbed for 30 seconds, then plunged into liquid ethane using a manual cryo-plunger in an ambient environment of 4 degrees C..
Chamber humidity: 50%
Chamber temperature: 277 K
Instrument: HOMEMADE PLUNGER
Details: Sample containing HIV STC intasomes in SEC buffer was applied onto freshly plasma-treated (6 seconds, Gatan Solarus plasma cleaner) holey gold UltrAuFoil grids (Quantifoil), adsorbed for 30 seconds, then plunged into liquid ethane using a manual cryo-plunger in an ambient environment of 4 degrees C..
Microscope: FEI TITAN KRIOS
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 300 kV
C2 aperture diameter: 100.0 µm
Nominal CS: 2.7 mm
Calibrated defocus: 1.5 µm - 4.0 µm
Nominal magnification: 22500.0
Calibrated magnification: 38167.0
Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER
Cooling holder cryogen: NITROGEN
Alignment procedure: COMA FREE
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 300 kV
C2 aperture diameter: 100.0 µm
Nominal CS: 2.7 mm
Calibrated defocus: 1.5 µm - 4.0 µm
Nominal magnification: 22500.0
Calibrated magnification: 38167.0
Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER
Cooling holder cryogen: NITROGEN
Alignment procedure: COMA FREE
Temperature
Minimum: 90.0
K
Maximum: 90.0 K
Maximum: 90.0 K
Image Recording
[1]
Detector model:
GATAN K2 SUMMIT (4k x 4k)
Detector mode: COUNTING
Dimensions: 3838 pixel x 3710 pixel
Frames per image: 1-100
Number of grids: 1
Number of real images: 1225
Average exposure time: 20.0 s
Average electron dose per image: 95.0 e/Å2
Details: Individual frames were gain-corrected, aligned, and summed with the application of an exposure filter using MotionCor2, according to the nominal dose rate.
Detector mode: COUNTING
Dimensions: 3838 pixel x 3710 pixel
Frames per image: 1-100
Number of grids: 1
Number of real images: 1225
Average exposure time: 20.0 s
Average electron dose per image: 95.0 e/Å2
Details: Individual frames were gain-corrected, aligned, and summed with the application of an exposure filter using MotionCor2, according to the nominal dose rate.
Final
reconstruction
Resolution: 3.9
Å
(
BY AUTHOR)
Resolution method: FSC 0.143 CUT-OFF
Number of images used: 83766
Algorithm: FOURIER SPACE
Resolution method: FSC 0.143 CUT-OFF
Number of images used: 83766
Algorithm: FOURIER SPACE
⌯ Applied Symmetry
Point group:
C2
Software
[1]
| Name | Version | Details |
|---|---|---|
| FREALIGN | 9.11 | - |
Startup model
[1]
Type:
INSILICO MODEL
Insilico model: Common lines model using OptiMod
Details:An initial model was generated directly from the class averages using OptiMod.
Insilico model: Common lines model using OptiMod
Details:An initial model was generated directly from the class averages using OptiMod.
⦨ Initial angle
assignment
Type:
PROJECTION MATCHING
Angular sampling: 7.5 degrees
Details: Relion 3D classification, auto mode
Angular sampling: 7.5 degrees
Details: Relion 3D classification, auto mode
Software
[1]
| Name | Version | Details |
|---|---|---|
| RELION | 1.3 | - |
⦩ Final angle assignment
Type:
PROJECTION MATCHING
Details: Frealign 3D classification and refinement
Details: Frealign 3D classification and refinement
Software
[1]
| Name | Version | Details |
|---|---|---|
| FREALIGN | 9.11 | - |
Particle selection
[1]
| Selected | Ref. model | Method | Software | Details |
|---|---|---|---|---|
| 274764 | - | - | - | - |
Final 3D classification
Software
[1]
| Name | Version | Details |
|---|---|---|
| FREALIGN | 3.11 | - |
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: CryoEM reconstruction of a tetrameric HIV-1 strand transfer complex intasome
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: CryoEM reconstruction of a tetrameric HIV-1 strand transfer complex intasome
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 192 | 192 | 192 |
| Origin | 0 | 0 | 0 |
| Spacing | 192 | 192 | 192 |
| Voxel size | 1.31 Å | 1.31 Å | 1.31 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 0.08 | AUTHOR |
