EMD-3847
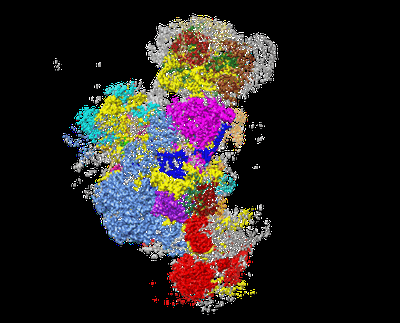
Cryo-EM structure of the 90S pre-ribosome from Chaetomium thermophilum
EMD-3847
Single-particle3.2 Å
 Deposition: 12/08/2017
Deposition: 12/08/2017Map released: 11/10/2017
Last modified: 25/11/2020
Name: The 90S Pre-ribosomne
Summary:
- Complex
- Protein
- Periodic tryptophan protein 2-like protein
- Utp2
- Utp3
- Utp4
- Utp6
- Utp7
- Utp10
- U3 small nucleolar RNA-associated protein 11
- Utp12
- Utp13
- Utp14
- Utp15
- Utp17
- Utp18
- Putative U3 snoRNP protein
- Utp24
- Utp30
- Nop1
- Putative nucleolar protein
- Nop58
- Snu13
- Rrp9
- RNA 3'-terminal phosphate cyclase-like protein
- Bms1
- Imp3
- Putative U3 small nucleolar ribonucleoprotein
- Putative U3 small nucleolar ribonucleoprotein protein
- Sof1
- Emg1
- KRR1 small subunit processome component
- Pre-rRNA-processing protein PNO1
- RNA cytidine acetyltransferase
- Fcf2
- 40S ribosomal protein S1
- 40S ribosomal protein S4
- 40S ribosomal protein s5-like protein
- 40S ribosomal protein S6
- 40S ribosomal protein S7-like protein
- 40S ribosomal protein S8
- 40S ribosomal protein s9-like protein
- 40S ribosomal protein S13-like protein
- 40S ribosomal protein S14-like protein
- 40S ribosomal protein S16-like protein
- 40S ribosomal protein S11-like protein
- 40S ribosomal protein S22-like protein
- 40S ribosomal protein s23-like protein
- 40S ribosomal protein S24
- 40S ribosomal protein S28-like protein
- Faf1
- RNA
- Ligand
Name: Periodic tryptophan protein 2-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [3]
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 100 kDa | - |
Name: Utp2
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 103 kDa | - |
Name: Utp3
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 73 kDa | - |
Name: Utp4
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 97 kDa | - |
Name: Utp6
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 46 kDa | - |
Name: Utp7
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 63 kDa | - |
Name: Utp10
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 198 kDa | - |
Name: U3 small nucleolar RNA-associated protein 11
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [3]
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 31 kDa | - |
Name: Utp12
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [3]
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 107 kDa | - |
Name: Utp13
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [3]
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 100 kDa | - |
Name: Utp14
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [1]
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 104 kDa | - |
Name: Utp15
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 60 kDa | - |
Name: Utp17
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 105 kDa | - |
Name: Utp18
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 69 kDa | - |
Name: Putative U3 snoRNP protein
Number of copies: 1
UniProtKB UniProtKB PDBe-KB PDBe-KB AlphaFold DB AlphaFold DB
Number of copies: 1
UniProtKB UniProtKB PDBe-KB PDBe-KB AlphaFold DB AlphaFold DB
Domains
Gene Ontology [2]
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 115 kDa | - |
Name: Utp24
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 22 kDa | - |
Name: Utp30
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 43 kDa | - |
Name: Nop1
Number of copies: 2
Number of copies: 2
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 32 kDa | - |
Name: Putative nucleolar protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 57 kDa | - |
Name: Nop58
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 64 kDa | - |
Name: Snu13
Number of copies: 2
Number of copies: 2
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 13 kDa | - |
Name: Rrp9
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 69 kDa | - |
Name: RNA 3'-terminal phosphate cyclase-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [4]
Biological process:
RNA processing ribosome biogenesis
Molecular function:
catalytic activity
Cellular location:
nucleolus
RNA processing ribosome biogenesis
Molecular function:
catalytic activity
Cellular location:
nucleolus
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 43 kDa | - |
Name: Bms1
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [11]
Biological process:
ribosome biogenesis regulation of rRNA processing endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)
Molecular function:
U3 snoRNA binding GTP binding GTPase activity
Cellular location:
cytoplasm small-subunit processome 90S preribosome nucleolus
ribosome biogenesis regulation of rRNA processing endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)
Molecular function:
U3 snoRNA binding GTP binding GTPase activity
Cellular location:
cytoplasm small-subunit processome 90S preribosome nucleolus
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 134 kDa | - |
Name: Imp3
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 21 kDa | - |
Name: Putative U3 small nucleolar ribonucleoprotein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 34 kDa | - |
Name: Putative U3 small nucleolar ribonucleoprotein protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [3]
Biological process:
rRNA processing
Cellular location:
Mpp10 complex sno(s)RNA-containing ribonucleoprotein complex
rRNA processing
Cellular location:
Mpp10 complex sno(s)RNA-containing ribonucleoprotein complex
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 86 kDa | - |
Name: Sof1
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 50 kDa | - |
Name: Emg1
Number of copies: 2
Number of copies: 2
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 27 kDa | - |
Name: KRR1 small subunit processome component
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [7]
Biological process:
rRNA processing endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)
Molecular function:
RNA binding
Cellular location:
ribonucleoprotein complex small-subunit processome preribosome, small subunit precursor nucleolus
rRNA processing endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)
Molecular function:
RNA binding
Cellular location:
ribonucleoprotein complex small-subunit processome preribosome, small subunit precursor nucleolus
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 37 kDa | - |
Name: Pre-rRNA-processing protein PNO1
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [4]
Biological process:
ribosome biogenesis
Molecular function:
RNA binding
Cellular location:
nucleolus cytoplasm
ribosome biogenesis
Molecular function:
RNA binding
Cellular location:
nucleolus cytoplasm
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 28 kDa | - |
Name: RNA cytidine acetyltransferase
EC number: 2.3.1.-
Number of copies: 2
UniProtKB PDBe-KB AlphaFold DB
EC number: 2.3.1.-
Number of copies: 2
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [12]
Biological process:
mRNA modification tRNA acetylation rRNA modification ribosomal small subunit biogenesis
Molecular function:
N-acetyltransferase activity snoRNA binding rRNA cytidine N-acetyltransferase activity tRNA binding ATP binding
Cellular location:
nucleolus preribosome, small subunit precursor small-subunit processome
mRNA modification tRNA acetylation rRNA modification ribosomal small subunit biogenesis
Molecular function:
N-acetyltransferase activity snoRNA binding rRNA cytidine N-acetyltransferase activity tRNA binding ATP binding
Cellular location:
nucleolus preribosome, small subunit precursor small-subunit processome
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 119 kDa | - |
Name: Fcf2
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 22 kDa | - |
Name: 40S ribosomal protein S1
Number of copies: 1
UniProtKB UniProtKB PDBe-KB PDBe-KB AlphaFold DB AlphaFold DB
Number of copies: 1
UniProtKB UniProtKB PDBe-KB PDBe-KB AlphaFold DB AlphaFold DB
Domains
Gene Ontology [3]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
cytosolic small ribosomal subunit
translation
Molecular function:
structural constituent of ribosome
Cellular location:
cytosolic small ribosomal subunit
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 29 kDa | - |
Name: 40S ribosomal protein S4
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [5]
Biological process:
translation
Molecular function:
rRNA binding structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
translation
Molecular function:
rRNA binding structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 29 kDa | - |
Name: 40S ribosomal protein s5-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [6]
Biological process:
rRNA export from nucleus translation
Molecular function:
RNA binding structural constituent of ribosome
Cellular location:
small ribosomal subunit cytosolic small ribosomal subunit
rRNA export from nucleus translation
Molecular function:
RNA binding structural constituent of ribosome
Cellular location:
small ribosomal subunit cytosolic small ribosomal subunit
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 23 kDa | - |
Name: 40S ribosomal protein S6
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [5]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex cytoplasm ribosome
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex cytoplasm ribosome
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 27 kDa | - |
Name: 40S ribosomal protein S7-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [4]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 23 kDa | - |
Name: 40S ribosomal protein S8
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [4]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 23 kDa | - |
Name: 40S ribosomal protein s9-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [4]
Biological process:
translation
Molecular function:
rRNA binding structural constituent of ribosome
Cellular location:
small ribosomal subunit
translation
Molecular function:
rRNA binding structural constituent of ribosome
Cellular location:
small ribosomal subunit
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 22 kDa | - |
Name: 40S ribosomal protein S13-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [7]
Biological process:
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) translation
Molecular function:
small ribosomal subunit rRNA binding structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome cytosolic small ribosomal subunit
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) translation
Molecular function:
small ribosomal subunit rRNA binding structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome cytosolic small ribosomal subunit
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 16 kDa | - |
Name: 40S ribosomal protein S14-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [5]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome cytoplasm
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome cytoplasm
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 16 kDa | - |
Name: 40S ribosomal protein S16-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [5]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex cytoplasm ribosome
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex cytoplasm ribosome
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 15 kDa | - |
Name: 40S ribosomal protein S11-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [4]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 18 kDa | - |
Name: 40S ribosomal protein S22-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [4]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 14 kDa | - |
Name: 40S ribosomal protein s23-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [3]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
small ribosomal subunit
translation
Molecular function:
structural constituent of ribosome
Cellular location:
small ribosomal subunit
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 15 kDa | - |
Name: 40S ribosomal protein S24
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [7]
Biological process:
methylation translation
Molecular function:
methyltransferase activity structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome cytoplasm
methylation translation
Molecular function:
methyltransferase activity structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome cytoplasm
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 15 kDa | - |
Name: 40S ribosomal protein S28-like protein
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Number of copies: 1
UniProtKB PDBe-KB AlphaFold DB
Domains
Gene Ontology [4]
Biological process:
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
translation
Molecular function:
structural constituent of ribosome
Cellular location:
ribonucleoprotein complex ribosome
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 7 kDa | - |
Name: Faf1
Number of copies: 1
Number of copies: 1
Natural source
Molecular weight
| Experimental | Theoretical | Method |
|---|---|---|
| - | 34 kDa | - |
