EMD-20133
Lon Protease from Yersinia pestis with Y2853 substrate
EMD-20133
Single-particle3.0 Å
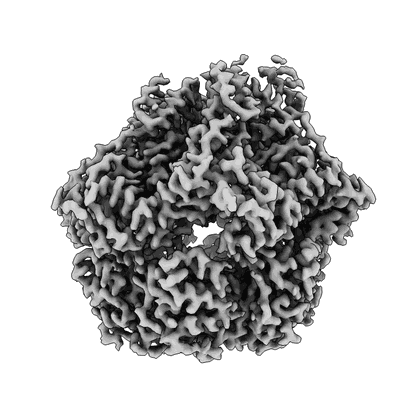 Deposition: 19/04/2019
Deposition: 19/04/2019Map released: 01/05/2019
Last modified: 20/03/2024
Concentration: 0.95
mg/mL
Details: This sample was monodisperse
Details: This sample was monodisperse
Buffer
pH: 8.0
Buffer components [5]:
Details: Solutions were made fresh from concentrated and filtered using a 0.1 um syringe filter to avoid microbial contamination. Buffers were stored on ice and used within 15 minutes of mixing in order to avoid excess ATP hydrolysis.
Buffer components [5]:
| Name | Formula | Concentration | ChEBI |
|---|---|---|---|
| Tris Base | Tris | 50.0 mM | |
| Potassium Chloride | KCl | 75.0 mM | |
| Magnesium Chloride | MgCl2 | 10.0 mM | |
| TCEP | TCEP | 1.0 mM | |
| Adenosine Triphosphate | ATP | 1.0 mM |
Grid
Mesh: 300
Model: Quantifoil, UltrAuFoil, R1.2/1.3
Material: GOLD
Details: Grids were plasma treated for 30 seconds using a 15 mA current operating under atmospheric gases using a glow discharger (Electron Microscopy Sciences).
Model: Quantifoil, UltrAuFoil, R1.2/1.3
Material: GOLD
Details: Grids were plasma treated for 30 seconds using a 15 mA current operating under atmospheric gases using a glow discharger (Electron Microscopy Sciences).
Pretreatment
Vitrification
Cryogen name: ETHANE
Chamber humidity: 95%
Chamber temperature: 277 K
Instrument: HOMEMADE PLUNGER
Details: 4 uL of sample was applied per grid and manually blotted for 4 seconds followed by immediately plunge-freezing in liquid ethane cooled by liquid nitrogen..
Chamber humidity: 95%
Chamber temperature: 277 K
Instrument: HOMEMADE PLUNGER
Details: 4 uL of sample was applied per grid and manually blotted for 4 seconds followed by immediately plunge-freezing in liquid ethane cooled by liquid nitrogen..
Microscope: FEI TALOS ARCTICA
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 200 kV
C2 aperture diameter: 70.0 µm
Nominal CS: 2.7 mm
Nominal defocus: 0.8 µm - 1.2 µm
Calibrated defocus: 0.5 µm - 1.5 µm
Nominal magnification: 36000.0
Calibrated magnification: 43478.0
Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER
Cooling holder cryogen: NITROGEN
Alignment procedure: COMA FREE ( Residual tilt: 0.14 mrad)
Details: Coma-free alignment procedure from Herzik & Wu, Nature Methods (2017). Preliminary grid screening was performed manually prior to data collection.
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 200 kV
C2 aperture diameter: 70.0 µm
Nominal CS: 2.7 mm
Nominal defocus: 0.8 µm - 1.2 µm
Calibrated defocus: 0.5 µm - 1.5 µm
Nominal magnification: 36000.0
Calibrated magnification: 43478.0
Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER
Cooling holder cryogen: NITROGEN
Alignment procedure: COMA FREE ( Residual tilt: 0.14 mrad)
Details: Coma-free alignment procedure from Herzik & Wu, Nature Methods (2017). Preliminary grid screening was performed manually prior to data collection.
Temperature
Minimum: 80.0
K
Maximum: 90.0 K
Maximum: 90.0 K
Image Recording
[1]
Detector model:
GATAN K2 SUMMIT (4k x 4k)
Detector mode: COUNTING
Dimensions: 3710 pixel x 3838 pixel
Frames per image: 0-43
Number of grids: 2
Number of real images: 4071
Average exposure time: 11.0 s
Average electron dose per image: 52.0 e/Å2
Details: Images were collected in counting mode at 4 frames per second
Detector mode: COUNTING
Dimensions: 3710 pixel x 3838 pixel
Frames per image: 0-43
Number of grids: 2
Number of real images: 4071
Average exposure time: 11.0 s
Average electron dose per image: 52.0 e/Å2
Details: Images were collected in counting mode at 4 frames per second
Final
reconstruction
Resolution: 3.0
Å
(
BY AUTHOR)
Resolution method: FSC 0.143 CUT-OFF
Number of classed used: 1
Number of images used: 118143
Algorithm: BACK PROJECTION
Resolution method: FSC 0.143 CUT-OFF
Number of classed used: 1
Number of images used: 118143
Algorithm: BACK PROJECTION
⌯ Applied Symmetry
Point group:
C1
Software
[1]
| Name | Version | Details |
|---|---|---|
| RELION | 2.0b | RELION 2.0b was used to perform final reconstruction |
Startup model
[1]
⦨ Initial angle
assignment
Type:
MAXIMUM LIKELIHOOD
Software
[1]
| Name | Version | Details |
|---|---|---|
| RELION | 2.0b | RELION 2.0b was used to assign initial euler angles |
⦩ Final angle assignment
Type:
MAXIMUM LIKELIHOOD
Details: RELION 2.0b was used to assign initial angles
Details: RELION 2.0b was used to assign initial angles
Software
[1]
| Name | Version | Details |
|---|---|---|
| RELION | 2.0b | RELION 2.0b was used to assign final euler angles |
Particle selection
[1]
| Selected | Ref. model | Method | Software | Details |
|---|---|---|---|---|
| 1176206 | - | - | - | Template-based cross correlation with FindEM |
Final 3D classification
Number of classes:
4
Avg. number of members per classes: 75000.0
Details: The final 3D classification had a somewhat asymmetric distribution owing to preferred specimen orientation due to interactions with the air-water interface
Avg. number of members per classes: 75000.0
Details: The final 3D classification had a somewhat asymmetric distribution owing to preferred specimen orientation due to interactions with the air-water interface
Software
[1]
| Name | Version | Details |
|---|---|---|
| RELION | 2.0b | RELION 2.0b was used to perform final classification |
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: Composite stitched map for final model building and refinement
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: Composite stitched map for final model building and refinement
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 130 | 130 | 130 |
| Origin | 0 | 0 | 0 |
| Spacing | 130 | 130 | 130 |
| Voxel size | 1.15 Å | 1.15 Å | 1.15 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 0.0403 | AUTHOR |
