Sequence search on a structural database is a major challenge. There are many tools that perform fast sequence matching, however, few support ProSite patterns and most are optimized for long sequences and are not effective for a short one.
This search is going as addition to ligand environment search and can be combined with the first one. The pattern it self is sequence motif which characterises properties of a protein chain. Matching and searching this motifs can bridng a new content to the familiar PDB entries.
In MSDsite search interface the pattern can be specified in terms of ProSite code or in compliant form with ProSite pattern motif.
Motif example:
A small sequence can be specified instead of a pattern. The only demand is that residues have to be separated by dash(-). Thus the search by a small sequence can reveal its binding properties.
Consider questions that this search is intended to answer we provide 2 examples for each one of them and 1 complex example.
Browsing results of ligand search from the "Ligand search"
chapter ot the tutorial where ligand is anticancer
drug residue MTX, we can find that the interacting ProSite
pattern is
PS00075.
Use the link for more information about this pattern.
In short it is dihydrofolate reductase signature.
motif:[LVAGC]-[LIF]-G-x(4)-[LIVMF]-P-W-x(4,5)-[DE]-x(3)-[FYIV]-x(3)-[STIQ]
Now we can explore more about proteins which sequence match the pattern.

The input should look like this:
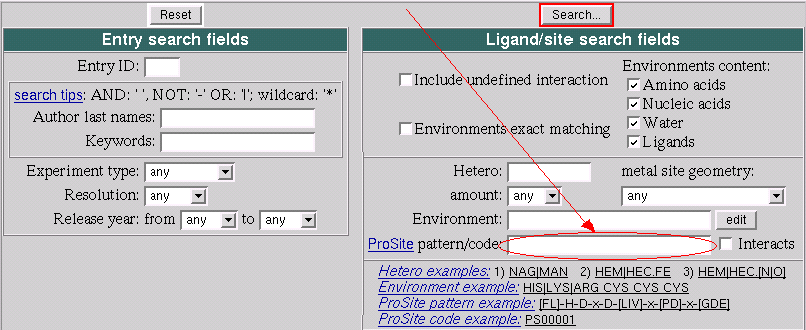
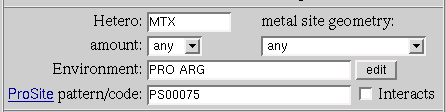
Assume that we want to invent a pattern which is specific for the such ligand type as MTX or similar ligands. In general it is rather complex task which demands a lot of work to be done. So here we consider a simplified version to demostrate help that a researcher can get from MSDsite.
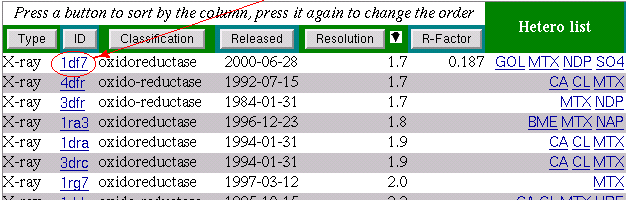
The part of interest starts after 40 sequence
number.Within this part of the sequence the MTX interacts with
the two protein residues Proline and Argenine.
We select the part of the sequence starting one residue
before the Proline and finishing at the Argenine:
LPAKVRPLPGR.
We do it using mouse with the pressed left button.
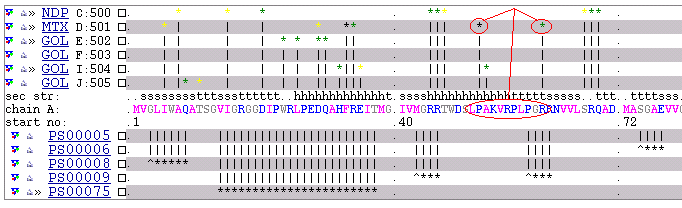
Now we have small sequence of interest: LPAKVRPLPGR
to be able to search by this sequence using MSDsite we
have to transform it into the comliant with the pattern format form:
L-P-A-K-V-R-P-L-P-G-R
At this stage we perform the search by this pattern. To do it
we put our pattern into start search from "ProSite
pattern/code"
field and press "Search..." button.

The result shows that the pattern is rather strict and was
found just in a few PDB entries.
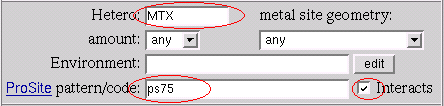
The search result brings us the 6 PDB entries: 1df7, 1dra, 3cd2, 3dfr, 3drc, 4dfr
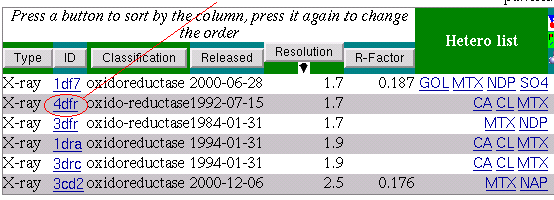

MTX * * * * 1df7: L-P-A-K-V-R-P-L-P-G-R 1dra: W-E-S-I-G-R-P-L-P-G-R 3cd2: I-P-L-Q-F-R-P-L-K-G-R 3dfr: E-S-F-P-K-R-P-L-P-E-R 3drc: W-E-S-I-G-R-P-L-P-G-R 4dfr: W-E-S-I-G-R-P-L-P-G-R
[LIWE]-[PES]-x(3)-R-P-L-[PK]-[GE]-R
For better understanding the binding properties of the pattern we use "ProSite pattern statistics" page.
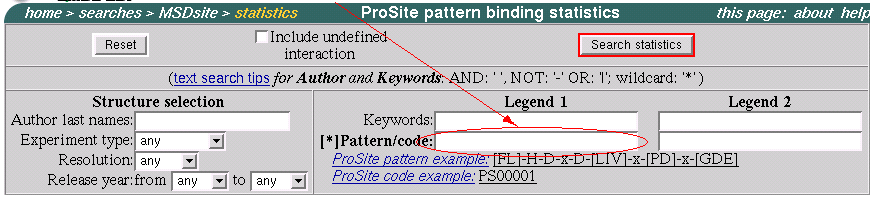
MTX is a favorite ligand of this pattern in sense of quantity. Analyzing the charts we see that most of ligands that interact with the new found pattern have a very similar to MTX structure. For example: from the table in the bottom of the page we can see that FOL/DDF/FA (Folic acids) 99% interact with the pattern. This is a sign of a quality of the pattern.