The interactive statistics page provides a way to view a statistical information regarding the interaction of a ligand with respect to residues that form an environment. The environment is a set of neighbour residues that interact with the ligand through a number of interaction types. It is possible to view two sets of ligand information on the same returned graph to allow comparison of the environment of these ligands.
Here we classify a residue type as amino-acid, nucleic-acid, water and ligand. This classification is parameter of distribution. The secondary structure can be specified as distribution parameter as well and it can be combined with the residue type.
Description of a ligand follows the rules from the ligand search and has the same flexible format.
To start search ligand statistics go to the start page of MSDsite (
http://www.ebi.ac.uk/msd-srv/msdsite and click on the line
"Ligand statistics" that is in the left top coner of the page as
shown on the picture below:
Let's consider particular questions that we can answer using this service.
After this modifications the form should look like: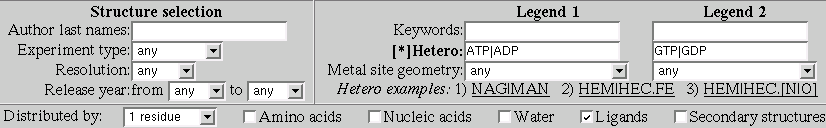
As we can see the undoubted leader is Magnesium (MG).
Now we can question our self: which site of ATP/ADP/GTP/GDP is interacting with MG?
This will bring us a chart of atomic bonds distrigution between ATP|ADP on the one side and MG on the other.
As can be seend from the chart the only phosphade part of ATP/ADP is involved in the interactions.
Going back (back button in the browser) to the ATP|ADP/GTP|GDP interactions chart and by clicking on the bar that corresponds to interactions between GTP/GDP and MG we are getting pretty much the same atomic bonds distribution that reveals that GTP|GDP interacts with MG by their phosphade part and all interactions are MG - Oxygen.
To answer the question we set up the statistics form as shown
on the picture: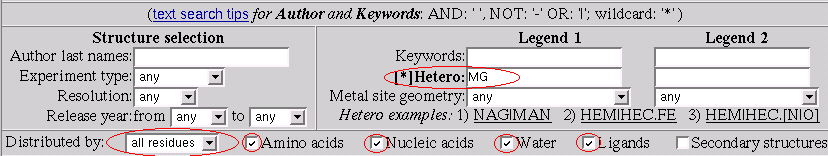
Now by click on the "Search statistics" button we get back the chart that is answer to the question above.
Let's explore this chart little bit more consider the
environment:
GLU GLU HIS HOH HOH HOH
GLU GLU HIS HOH HOH HOHAs a respond the service brings back a list of PDB entries that have MG within this environment.
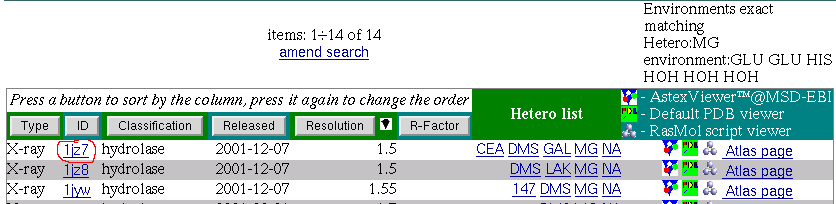
 if you have a Rasmol script application setted up for mime-type:
application-x/rasmol or go to the link under image
if you have a Rasmol script application setted up for mime-type:
application-x/rasmol or go to the link under image
 to load the structure in the viewer applet.
Rasmol gives a picture something like below:
to load the structure in the viewer applet.
Rasmol gives a picture something like below: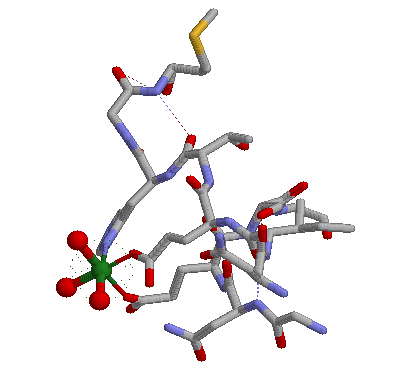
Here we can see MG octahedral metal coordination formed by three water molecules, two Glutamic acids and one Histidine.
Using MSDchem service we can obtain necessary information about ligands that contain these groups and their atoms names.
Adenosine is contained in ATP,ADP,A chemical compounds and is described by atoms N1,N3,N6,N7,N9,C2,C8 (other carbons are well hidden and not active).
Guanozine is contained in GTP,GDP,G chemical compounds and is described by atoms N2,N1,N3,N7,N9,O6,C8 (other carbons are well hidden and not active).
ATP|ADP|A.N6|N1|N3|N7|N9|C2|C8GTP|GDP|G.N2|N1|N3|N7|N9|O6|C8Now let's explore the interactions in more details.
this brings the atomic bonds distribution between Adenosine and Glutamic acid
As can be seen from this chart Adenosines atom N6 is the absolute leader in interactions with Glutamic acid and most interactions are Hydrogen bonds. Moreover a lot of interactions are with Glutamic main chain atom O.
this brings the atomic bonds distribution between Guanozine and Glutamic acid
Let's find out which site of Guanozine is most biologically active.
Now let's consider what is happening in 3D
This will bring us a list of PDB entries that correspond to hydrogen bond interactions between GDP.N1 and ASP.OD1
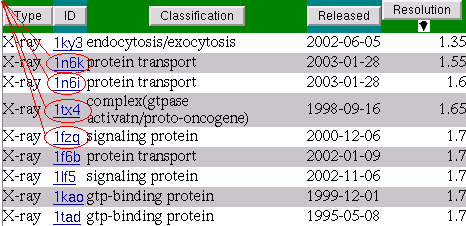
Let's go into detail pages of these PDB entries starting from the first one. We will add all matched ligands from these detail pages to the Multi-View page.
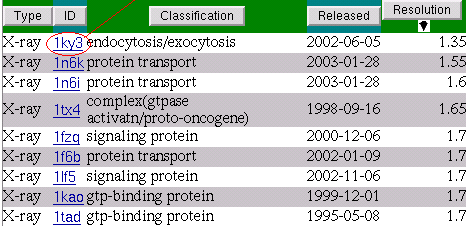
The detail page presents interactions of the ligands on the residual level.
Now we are going to add the ligand to the Multi-View page for comparison of binding properties in 3D.
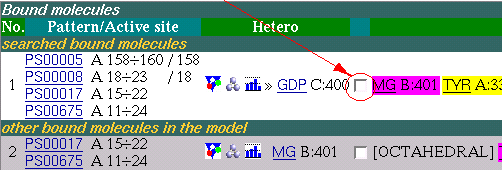
Note:
If you've done it for the first time during the
session then a new browser window will be opened,
otherwise the information about the ligand occures in
the existing window.
Now let's have a look what we supposed to have on the
Multi-View page: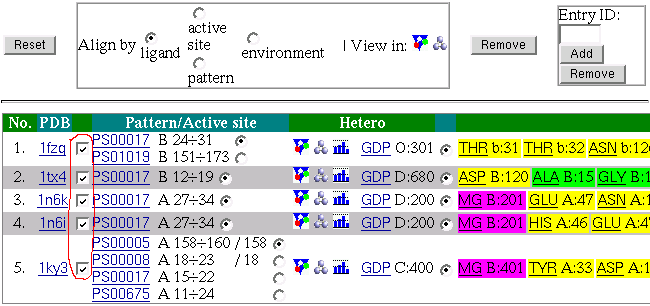
 if you have a Rasmol script application setted up for mime-type:
application-x/rasmol or go to the link under image
if you have a Rasmol script application setted up for mime-type:
application-x/rasmol or go to the link under image
 to load the structure in the viewer applet.
Rasmol gives a picture something like below:
to load the structure in the viewer applet.
Rasmol gives a picture something like below:
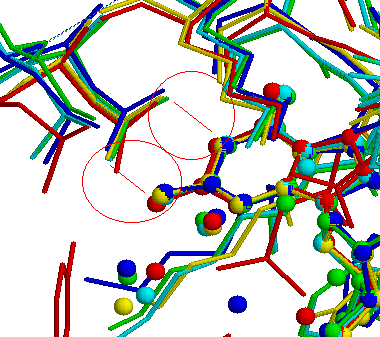
After zooming the view we get a more clear picture about what is
happening between Guanozine atoms N1,N2 and Aspatic acid atoms
OD1,OD2. It is bidentide Hydrogen bond interaction: