Ligand search
Here we consider search by a given ligand and/or its environment
This search is intended to answer the following questions:
- Which PDB entries contain a given ligand?
- Which PDB entries contain a given ligand interacting by a
specified site?
- Which PDB entries contain small molecules within the given
ligand environment?
- Which PDB entries contain metal within a given site
geometry.
- Combined question from the previous four
Ligands interaction with its environment can be described on
residue and atomic levels.
For extended search residue got attribute that characterize it
as belonging to a secondary structure and atom got attribute
that characterize it as belonging to an atoms group like
base, sugar, phosphide, guanine...
In addition to the environment description for metal sites the
desired site geometry can be specified as well.
For this type of search a small molecule and/or its environment
and/or metal site geometry have to be specified.
For residue or small molecule type
identification we are using its three letters code. Atoms name
are following PDB convention. Residue name and atom name are
separated by dot. It is available to specify general element
instead of atom name. In this case it has to be closed in square
brackest.
To make life easier the graphical query interface is provided
to setup ligand environment where in a convenient manner all
mentioned above search attributes can be specified. The search
templates can be stored in a separate file and used in another
session.
Consider questions that this search is intended to answer we
provide four examples.
Which PDB entries contain the anticancer drug residue MTX?
Steps:
- In your browser go to: http://www.ebi.ac.uk/msd-srv/msdsite/
- Type MTX in the "Hetero" field of the search
form.
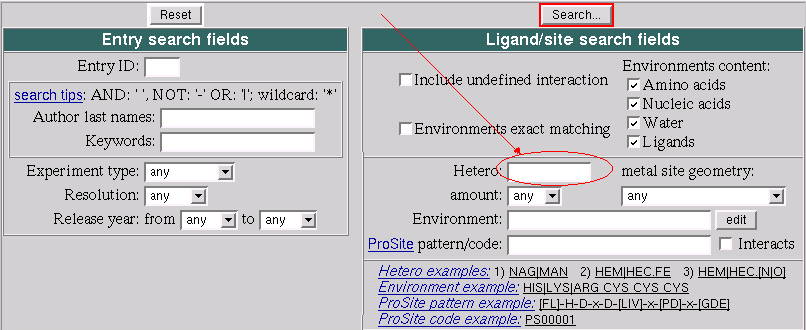
- Click "Search..." button and get back a list of the PDB
entries
Which PDB entries contain the anticancer drug residue MTX interacting by its Oxygens?
Steps:
- In your browser go to: http://www.ebi.ac.uk/msd-srv/msdsite/
- Type MTX.[O] in the "Hetero" field of the search form.

- Click "Search..." button and get back a list of the PDB
entries
Which PDB entries contain small molecules within next environment?
Note: Residues of environment are somewhere around a ligand. They
interact with a ligand but they are not in a sequence.
Here we specify the environment which is common for the
anticancer drug residue MTX
List of the environment residues
- Aspatic or Glutamic
- Isoleucine or Leucine or Valine
- Argenine or Lysine
- Phenalalanine or Proline or Triptophan
To perform the search follow the steps:
- In your browser go to: http://www.ebi.ac.uk/msd-srv/msdsite/
- Type ASP|GLU ILE|LEU|VAL ARG|LYS PHE|TRP|PRO in the "Environment" field of the search form.
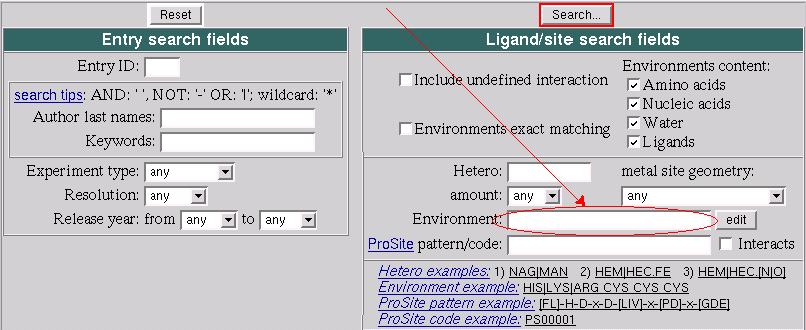
- Click "Search..." button and get back a list of the PDB
entries
Combined search by ligand and it's environment
For this example the result from the previous one can be
used for the start.
Assume that we look at the result page from the previous
example. There is a link on the top of this page: amend
search.
- Go to the link amend search.
As the result the search conditions are displayed and
"Environment" filed contains ASP|GLU ILE|LEU|VAL ARG|LYS
PHE|TRP|PRO.
- Click edit button that is to the right from
the "Environment" field.
This will bring you to the "graphical query generator" by
Dr. Tom Oldfield MSD:EBI-EMBL.
The editor window contains empty oval shape that is
associated with a ligand and four rectangles which are
associated with the environment residues.
- In the window of this tool point a mouse to the oval shape that is
associated with a ligand then press left shift button on your
keyboard and left click mouse.
The dialog associated with the ligand has
appeared.
- In the appeared "Ligand" dialog box
- type MTX in the "Code 3 letter" field
- choose "General element" radio button
- type O for Oxygen in the associated with the atom
second fied
- click "Ok" button
- Now drag and drop 3 boxes (with ASP,GLU with ILE,LEU,VAL and
with PHE,TRP,PRO ) one by one to the "Trash
bin". So the box with ARG,LYS has left.
- Now move the mouse to point it to the circle between the ligand
(MTX) and the interacting residue (ARG,LYS) then hold the left
shift button on the keyboard and left click mouse.
The dialog box with the interaction parameters has
appeared.
- In the "Interaction" dialog box choose "Bond length" as
"Less than" and drag the slide to set up distance equal
3.0
- Close "Interaction" dialog box by clicking on "Ok"
button.
- Click "Submit" button in the editor panel
This way you've specified the next query to MSD:
Find all PDB entries where anticancer drug residue MTX
interacts by its Oxygens with a protein residue which is
Arginine or Lysine and interaction distance is no more
then 3 Angstroms.
- Click "Search..." button and get the PDB entries list.



