
The Protein Data Bank contains a significant number of protein structures that have ligands bound. One of the most important uses of macromolecular structure data is the study of the interactions between macromolecules and bound ligands. Due to the local nature of the interactions between ligands and macromolecules, ligand binding sites are often more highly conserved across a functional family than the overall structure and fold of the macromolecule. As such, a catalogue of ligand-macromolecule interactions can be invaluable in the wider study of structure-function relationships. The specific three-dimensional environments of the protein binding sites for different ligands has been derived from the parsing and loading of the PDB entries into a relational database. Binding site descriptors have been derived and a powerful search and retrieval system has been developed.
MSDsite is the database and its query interface located at: http://www.ebi.ac.uk/msd-srv/msdsite.
This project is funded by the EU TEMBLOR award.
Other non commercial protein-ligand search engines:
- Relibase: http://relibase.ebi.ac.uk
- IMB Jena Image Library of Biological Macromolecules http://www.imb-jena.de/IMAGE.html
- Goto, S., Okuno, Y., Hattori, M., Nishioka, T. and Kanehisa, M.; LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res. 30, 402-404 (2002).
- V. Santander, M.A. Portales and F. Melo .(2003) A tool to assist the study of specific features at protein binding sites Bioinformatics 19, 250-251 http://protein.bio.puc.cl
- PDBSite - a database on protein active sites and their spatial environment
Ivanisenko V.A., Grigorovich D.A., Kolchanov N.A. PDBSite: a database on biologically active sites and their spatial surroundings in proteins with known tertiary structure. The Second International Conference on Bioinformatics of Genome Regulation and Structure (BGRS'2000), Novosibirsk, Russia, August 7 - 11, 2000, Vol. 2, pp. 171-174.
http://wwwmgs.bionet.nsc.ru/mgs/gnw/pdbsite/
- PROMISE. The Prosthetic groups and Metal Ions in Protein Active Sites Database
Degtyarenko, K.N., North, A.C.T. and Findlay, J.B.C. (1999) PROMISE: a database of bioinorganic motifs. Nucleic Acids Res. 27, 233-236. http://metallo.scripps.edu/PROMISE/
- Castagnetto, J.M.; Hennessy, S.W.; Roberts, V.A.; Getzoff, E.D.; Tainer, J.A.; Pique, M.E. "MDB: the Metalloprotein Database and Browser at The Scripps Research Institute". Nucleic Acids Res. 2002, 30, 379-382.
http://metallo.scripps.edu/
Database
MSDsite data is part of PDBe and consists of:
- Ligands and their interactions with macromolecules
(proteins/RNA/DNA), with other ligands and with solvents
(water).
- Geometries of metal sites
- Sequences and ProSite patterns with their ligand interactions.
- Bound molecule coordinates
The bound molecules are small chains of hetero groups (one or
more) that are covalently bonded. All PDB entries are
analyzed and information about bound molecules are generated
based on this rule.
Information about hetero groups is stored in the reference
database that is part of MSD.
Data
MSDsite allows access to the ligand environment
in a particular PDB entry and contains coordinates with the
respect to this entry. The derived bound molecule environments
include a consideration of the symmetry in PDB X-ray entries.
All possible symmetry and translations are considered to
list all possible protein ligand contacts. Water coordinates
are treated in a similar manner.
Bonds
The Inter atomic interactions stored in PDBe have the classifications:
- Covalent bond
- Ionic bond
For metal atom interactions the
classification as either a covalent or ionic bond has been considered
as a probability between the likelihood as whether metal atom
interacts more like covalent or ionic coordination using the following:
For the elements: F Cl Br I At :
- If the atom is a PDB HETNAM chemical group, and
is not an ion, it can form one covalent bond.
- If the atom is a PDB HETNAM chemical group, and
is an ion, it cannot form covalent bonds.
- If the atom is a member of a PDB HETNAM group that contains
other atoms, it cannot form covalent bonds with
any atom of another group.
The following metals are not expected to form
bonds to carbon but are allowed O and N coordination, except
for organometallic clusters and carbon monoxide:
Li Na K Rb Cs Fr Be Mg Ca St Ba Ra La Ce Pr Nd Pm Sm Eu Gd Tb Dy Ho Er Tm Yb Ac Th Pa U
The following elements may form bonds to
C and S : As Sb Bi Sn Pb Al Ga In Tl
For the following elements a rule of thumb
(see below) was used to distinguish between covalent and
ionic interactions:
Zn Cd Hg Sc Y Lu
Rule of thumb:
- If distance between two atoms is within
covalent bond length then bond is covalent
coordination.
- If distance between two atoms is within (ionic
+ covalent) / 2 then bond is covalent
coordination.
- Otherwise if distance is within ionic bond
length then it is ionic coordination bond.
For the following elements, the
rule of thumb was applied, except for Carbon which was always
considered as forming a covalent coordination bond: Ti V Cr Mn Fe Co Ni Zr Nb Mo Tc Ru Rh Pd Ag Hf Ta W Re Os Ir Pt Au.
B is always covalent in PDB.
- Disulfide bond
- Hydrogen bond
We consider two types of Hydrogen bonds based on
an algorithm/approach.
- Hydrogen bonds are calculated using
hybridization, energy type and binding geometry of
potential donor-acceptor pairby the methods
described in HBExplore.
- Hydrogen bonds that are calculated using bndlist
library.
Hydrogen bond assignment is
further processed where if the first approach is used and
if it did not recognize a pair of atoms as potential
donor-acceptor
then the second algorithm is applied.
- Van Der Waals bond
- Planes
We consider two types of plane, planar chemical entities and all rings that satisfy a set of rules are
treated as planar groups.
- Merged full set of smallest rings.
For the rings set the definition
from Oelib is used
(build a minimal spanning tree).
Each residue is examined in terms of
containing rings by first finding the full set of minimal rings.
The rings having two or more atoms in common are merged. The
result is always the desired ring itself.
A plane is built from the rings found according
to the following rules:
-
Validate ring
- If ring has 8 atoms or more then it is eliminated
- If ring has elements B P S then
it is eliminated
- At this stage we assume that the rings are
valid and we cleanup the set of atoms that define
the rings set to form a plane.
- Eliminate bridge atoms
- Eliminate metal atoms
- If plane has more then three atoms then store it in MSD.
- Planar chemical entities or sub-structures of 4 and more
atoms.
For example the carboxylate end of a glutamic acid side chain.
To define such sub-structures Vega atom topology typing was used
The following rules are applied:
- Plane must consist of C-3#0, N-2#0, O-1#0 atoms
with Vega type, as a terminate C-4#0, N-300,
O-2#0 are appropriate, all the other atoms are
eliminated. C-3#0 means that general element is
Carbon, the atom is covalently bonded to other 3
atoms in the residue, it can be in a ring. The
last one digit says whether the ring is aromatic
or not, if it is 0 then aromatic ring is not
appropriate (planes from aromatic rings was
built on the step one - planes from rings, see
above).
- Atoms must be connected and be in a
geometrical plane, where atom deviations from the plane are
within 0.1 Angstroms.
- The number of atoms in a plane must be greater than 3.
Plane-Plane interactions are stored where the distant between
the centres of gravity are within 6 Angstroms, the
the distance and normal vectors to the planes are
stored.
- Non bonding interactions
A Non bonding type is assigned to interaction between two
atoms that are within 4 Angstroms and which bond
type has not been classified.
The following figure shows relations between database tables for
storing interactions between small molecules and macromolecules:

Geometry
Metal coordination geometries are calculated and stored in MSD:
- tetrahedral
- triagonal bipyramidal
- square-based pyramidal
- octahedral and undefined
The metal sites were classified and geometry types
assigned based on all valid symmetry related ligand
positions. Metal site geometry definition angles
and distances and deviations from [6] are used.
Sequences and ProSite patterns
ProSite patterns were loaded into PDBe and a
matching algorithm was run for all PDB entriesand the matches
stored in the database.
For all non standard patterns (that are
not in ProSite list but comply to the ProSite pattern format) a
fast search algorithm was developed and embedded into the Oracle
database.
Search concept
The MSDsite search interface starts from the search form and
once a result is found one can then amend the search and get a family of similar PDB
entries. So each detail page about a particular PDB entry is at the
same time a search form. Even statistics pages with the
interactive charts are search forms themselves. Each result set
can be amended and to cope with this challenge the front page was
developed in such a way that any question, that can be asked from
statistics pages or from detail pages, can also be specified on the front page form to
get the same answer.
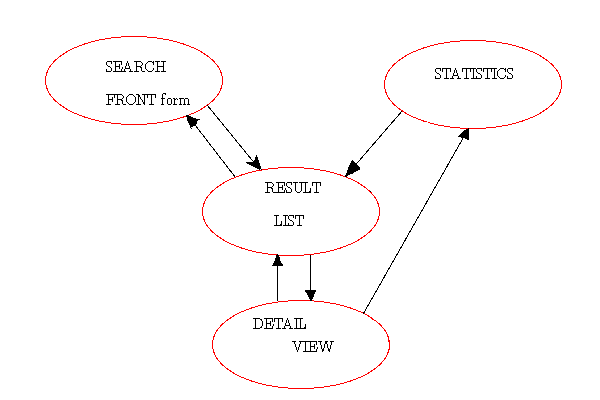
The second search concept is that search and statistics forms
have two parts: ligand search fields and structure filter. The
structure filter allows one to restrict a search set by PDB
header information like titles, keywords, header class,
resolution, release year, author.
Presentation concept
Now we consider user interface. What can be
presented and what are the relative weights of these items? From
the start internet was very sensitive to the data volume, but
with the high-tech revolution 1995 - 1996 we can benefit very fast
internet connection, approximately the same speed as
intranet. Keeping in mind that probably most users of MSDsite
are academic institutes which have fast internet access and not
always have enough calculations power we can include a lot of
pregenerated images in the web interface without slowing it
down. The ligands presentation needs 3D graphics and applets
could help here, but on the other hand their usage becomes more
and more restrictive. Most of the modern browsers come without
java, which has to be installed separately. The only solution is
to support several 3D visualization tools and MSDsite provides
Rasmol [10] script output and EBI-Astex Viewer [11]
applet. Summarizing MSDsite interface presents most popular in
web world html tables with lots of pop-up images and with links
to several 3D visualization tools.
In MSDsite we consider the two kinds of search result presentation:
- Interactive sortable list of PDB entries
- Interactive statistics char
- Interactive list of ligands from different PDB entries
In more details:
Interactive sortable list of PDB entries
The list presents description of PDB entries and has next functionality:
- Link to details pages for an entry
- Sequence view
- presents parts of chain sequence where it matches ProSite patterns or searched pattern or interacts with a ligand.
- Ligands view
- presents ligands and interaction with their environment. ProSite or searched pattern is presented as well where matched part of the sequence interacts with a ligand.
- Atomic interaction view
- presents interaction on atomic level- bond type,
distance, angle between planes and angles for a
single atom ligand.
- Links to 3D visualization tools
- Link to integrated resource - atlas pages (MSDlite, MSDPro)
- List can be listed and sorted interactively.
Interactive statistics charts
- Ligand binding statistics
- It provides a way to view statistical information
regarding the interaction of particular ligand(s) with
respect to residues that form an active site
environment. The environment is a set of neighbour residues
that interact with the ligand through a number of
interaction types. It is possible to view two sets of ligand
information on the same returned graph to allow comparison
of the environment of these ligands.
- Environment binding statistics
- It provides a way to view statistical information
regarding the environment of a ligand with respect to
ligands. It is therefore possible to define queries based on
residue type and find which ligands are bound to this
environment.
- Atomic bonds statistics
- A search is available to present and compare statistical
information for a pair ligand - macromolecule residue
interaction on atomic level.
- Sequence patterns binding statistics
- It provides a way to search PDBe for statistical
information regarding the pattern interaction with a ligand
with respect to ligands. It is therefore possible to define
queries based on pattern and find which ligands are bound to
this pattern.
Interactive list of ligands from different PDB entries
The order of this page is to provide functionality for 3D
alignment of ligands with their environment. Detailed pages
for a PDB entry have links to this page so any ligand can be
submitted here and then its geometry, environment and
interacting patterns can be compared with another one.
The interface has four alignment options:
- Alignment by ligand
- Alignment by environment
- Alignment by pattern
- Alignment by active-site residues



