
I choose a Zinc binding pattern PS00903.
PS00903 - Cytidine and deoxycytidylate deaminases zinc-binding region signature
[CH]-[AGV]-E-x(2)-[LIVMFGAT]-[LIVM]-x(17,33)-P-C-x(2,8)-C-x(3)-[LIVM]
In this example I consider 3D properties of the given PROSITE pattern.
I start with the pattern binding statistics : http://www.ebi.ac.uk/msd-srv/msdmotif/statisticspattern
I input PS00903 (PROSITE pattern code) on the form into the field
marked with the red circle: 
After that I click on "Search.." button in the top right
corner:
The service provides back an interactive chart as one shown below:

The chart presents statistics regarding ligand binding properties of this pattern throughout the PDB archive. Chart shows that ZN is the most often ligand in these interactions. I click now on the bar which corresponds to ZN and get back a hit list like on the picture below:

The hit list contains fragments of sequences from the PDB archive which match the pattern and where this pattern binds ZN.
The screen presents multiple alignment based on the pattern
definition:
([CH]-[AGV]-E-x(2)-[LIVMFGAT]-[LIVM]-x(17,33)-P-C-x(2,8)-C-x(3)-[LIVM]).
This pattern has two flexible gaps : the first one from 17 to 33
residues long and the second one from 2 to 8 residues
long. Though as can be seen from the picture only the
first gap is varied between structures in the PDB. The
second gap between two Zinc binding Cystines is fixed to
be two residues long.
The interacting residues are colored in according with the type of ligand interaction:
The page provides sequences from the hit list in CLUSTALW format with the corresponding mime-type. A browser can be setup to use an application for further research of sequences. Here I use Jalview as browser helper application. To start it from the page I click on the link in the red circle. If you do it with me you might have a picture on your screen as provided below:

I leave here Jalview and return back to the MSDmotif hit list.

PS00903 pattern interacts with ZN by its first
residue and two Cystines at the end. This can be seen by
clicking on the image  in the red circle, which leads to the picture as below:
in the red circle, which leads to the picture as below:
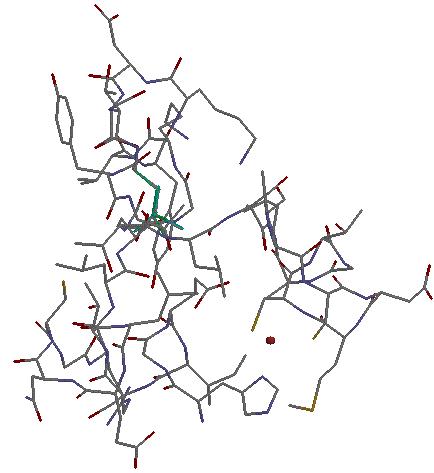
Returning back to the hit list:

It contains 43 hits from relatively the same PDB entries. To filter out similar PDB entries I click on "amend search" link and get the picture on the screen as below:

The blue circles present the initial search criteria: Pattern PS00903, Hetero: ZN and pattern interacts with the ligand.
In the right bottom corner of the form there is "Normalize by" drop down list where I choose Sequence 50%, which means that after the search is performed the PDB chains with more than 50% sequence identity will be filtered out.
To apply this change I click on the "Search..." button in the top right corner of the form, which is in the red circle on the picture provided.
The normalized hit list contains 8 hits from PDB chains with no more than 50% of sequence identity:

To view 3D alignment of the hits from the hit-list I select (tick) all check boxes on the same line as the hits (in red circle on the picture below):

And after the selection is made, I click on the image  in the red circle and get a Rasmol picture. To make it
less messy I input the rasmol
commands:
in the red circle and get a Rasmol picture. To make it
less messy I input the rasmol
commands:
select protein cartoon on wireframe off select ZN spacefill
The result must look like the following picture:

The end