

Here I consider a Calcium binding loop and discuss how to apply tools provided by MSDmotif for researching this site. I'll be using pattern binding statistics, ligand binding statistics, number of searches, including PHI/PSI search.
Few more words about PHI/PSI search. Phi/Psi parameters of the backbone can be translated into geometrical characteristics and vice versa. This way phi/psi gives a sequential representation of the proteins geometry. Sequential search prevents 3D search from exponential dependency on the number of residues making it linear. Thus Phi/Psi search is ideal for short fragments with an unknown secondary structure (loops).
To start this search I go to MSDmotif front page : http://www.ebi.ac.uk/msd-srv/msdmotif, and input "1gci" in the "PDB code" field which is marked with the red circle on the picture below:

After that I click on the "Search..." button in the top right corner and get back the hit list like one shown below:
Here I click on the image  in the red circle, to view the structure in a PDB viewer
which setup in my browser as a helper application on
mime-type: application/x-rasmol, see the picture below
in the red circle, to view the structure in a PDB viewer
which setup in my browser as a helper application on
mime-type: application/x-rasmol, see the picture below
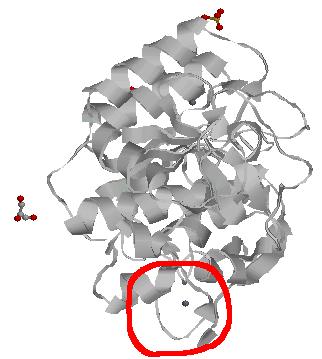
Shown on the picture is the 1gci PDB entry from the Subtilases SCOP family. The subject of this example is the Calcium binding loop in the red circle. This calcium binding site occur in Subtilases proteins. It could be interesting to find this loop in another proteins family.
I return to the hit-list with 1gci PDB entry in it and click on the link "1gci" in ID column. This link is in the red circle on the picture below.

The browser opens a MSDmotif details page with the respect of the given PDB entry (1gci):

The Calcium binding site is in the red circle. It is from the residue 75 to the residue 81 in the sequence. I have a look around to find out information about this look from the page.
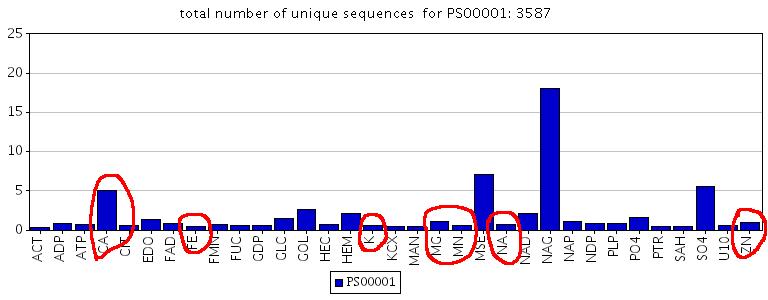
While looking at the detail page I can spot that there is one PROSITE pattern PS00001 inside this loop from the residue 76 to the residue 79. This is N-glycosylation site which quite often binds positively charged metals, especially Calcium. (see MSDmotif pattern statistics for PS00001)
The picture below highlights the pattern position in the sequence:

As MSDmotif pattern binding statistics witnesses (see http://www.ebi.ac.uk/msd-srv/msdmotif/barchartpattern?pattern1=PS00001), N-glycosylation site binds metals (see the picture below).
N-glycosylation site PROSITE pattern is defined as: N-{P}-[ST]-{P}
The first residue of the N-glycosylation site must be Asparagine (N - one letter code). Asparagine is similar to Aspartic (D - one letter code) but it has partial negative charge on the side chain:
| Asparagine: |
Aspartic: |
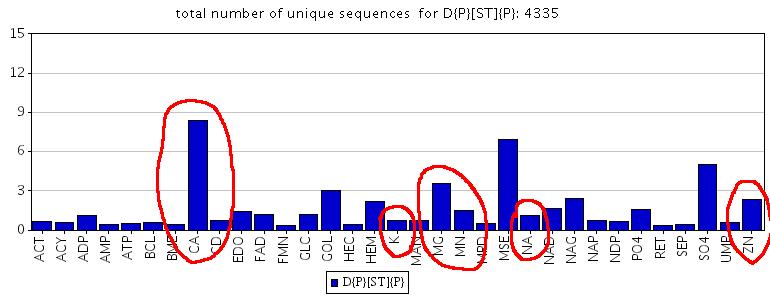
If I change Asparagine to Aspartic in the pattern (D-{P}-[ST]-{P}) and submit this request to the pattern binding statistics: http://www.ebi.ac.uk/msd-srv/msdmotif/barchartpattern?pattern1=D{P}[ST]{P} then I can see that the upgraded pattern is much more selective to metals, especially to Calcium, than the N-glycosylation site with Asparagine at the front (see the picture below):
Now I reverse the question from which ligands the given pattern binds to whether metals likely to bind N-glycosylation site or not. From the Periodic table I pick similar ligands to Calcium - metals: CA,K,MG,NA,SR. and submit them as a search criteria to the other MSDmotif statistics page: Ligand binding statistics. This gives me a picture like below:

I get the corresponding hits by clicking on the bar which corresponds to PS00001 PROSITE pattern and which is marked on the picture with the red circle.
Now my browser shows the hit list which looks like on the picture below:

There are many hits (914) most of them are from the same SCOP families. To see how many SCOP families are there in this list I click "amend search" link which is marked on the picture with the red circle.
Now on the front page of MSDmotif I change normalization drop down list to normalize by "SCOP family" as shown on the picture below:

I click on the "Search..." button in the top right corner and get hit list with 74 hits and consequently with 74 SCOP families.
Few more searches show that
About 18% of SCOP families which have N-glycosylation site and Calcium like metals in, have the N-glycosylation site interacting with Calcium like metals (CA,K,MG,NA,SR). Just for Calcium (not CA,K,MG,NA,SR, but just CA) this figure rises up to 22%.
About 20% of SCOP families which have D{P}[ST]{P} pattern and Calcium like metals in, have the D{P}[ST]{P} pattern interacting with Calcium like metals (CA,K,MG,NA,SR). Just for Calcium (not CA,K,MG,NA,SR, but just CA) this figure rises up to 28%.
I return back to the detail page which presents Calcium binding loop in 1gci PDB entry.

Another motif which is found inside the Calcium binding loop is asx-turn. It is highlighted with the red circle and red lines on the picture above. ASX-turn is stretched from the residue 77 to the residue 79.
This motif consists of three residues and starts with Aspartic (D one letter code) or Asparagine (N one letter code) amino-acids, for the definition of the motif visit the link above.
Now I consider how many SCOP families have this motif bound to the Calcium like metals?
To answer this question I open the http://www.ebi.ac.uk/msd-srv/msdmotif link to MSDmotif front page in my browser and click on the "go" link on the same line with "asx-turn" in the left bar of the form, this submits asx-turn to the search criteria. I tick "Interacts with a ligand" check box which is next to the motifs drop down list. Now I input CA|K|MG|NA|SR in the "Hetero" field of the form and select normalization by "SCOP family" from the drop down list in the right bottom corner of the form. My input looks like on the picture below where the input fields are marked with a red circle.

Now I click on the "Search..." button in the right top corner of the form to get the corresponding hit list as one shown on the picture below:

It has 145 hits from 145 SCOP families which is the answer to the question.
Few more searches show that about 25% of SCOP families which have asx_turn and Calcium like metals in, have a bond between asx_turn and Calcium like metals (CA,K,MG,NA,SR).
Just for Calcium (not CA,K,MG,NA,SR, but just CA) this figure rises up to 40%.
Now I'm interested in a 3D distribution of the metals around asx-turn motif.
I tick all check boxes in the same line as the hits (marked
with the red circle on the picture below) and click on the image  in the red circle, to view the 3D aligned structure fragments in a PDB viewer.
in the red circle, to view the 3D aligned structure fragments in a PDB viewer.

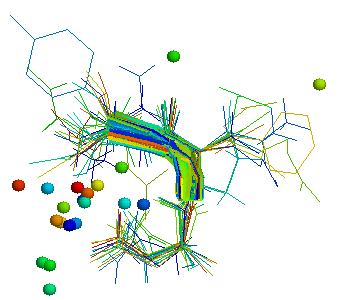
The picture above shows the concentration of the ligands inside the loop (asx-turn).
Here, again, I return back to the detail page which presents Calcium binding loop in 1gci PDB entry.

Now I abstract from the asx-turn motif definition and leave phi/psi angles only. Thus I remove the constraint that the first residue must be Aspartic or Asparagine.
I specify 77,79 in the "from" "to" fields of the from and click on the "Search.." button
Search brings 48 hits most of them from the same SCOP family as the original PDB entry 1gci - Subtilases (see the picture below)

To change the search criteria and to normalize the hit list by SCOP family I use "amend search"link which is marked by the red circle on the picture.
After I click on the "amend search"link the browsers shows me the MSDmotif from page with some field filled. The "sequence" field contains xxx, the "phi/psi" fields contain phi/psi angles for the given three residues from the structure. The "Phi/Psi deviation" field is set to 10 degrees (means that phi/psi angles can variate from the given +-10 degrees).
The following picture presents the original search criteria marked with the blue circles

At this point I change the search criteria:
Then I click on the "Search..." button and wait till the result come back...
The hit list consists of 14 hits from different SCOP families:
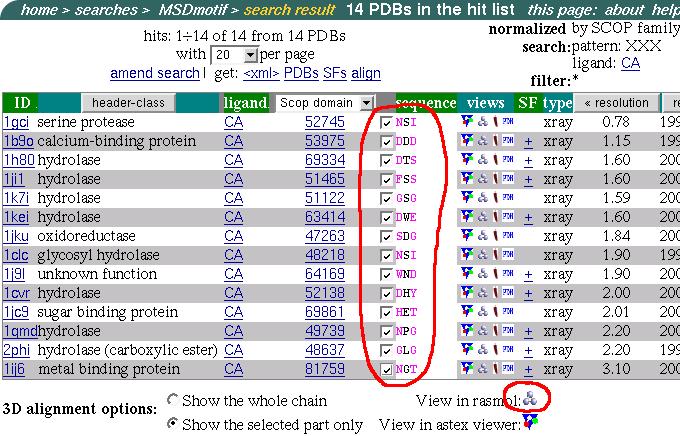
As can be seen there are other residues beside Aspartic (D) and Asparagin (N) presented in the list as a first residue of the motif. It is interesting to see how good they align in 3D.
I tick all check boxes in the same line as the hits (marked
with the red circle on the picture below) and click on the image  in the red circle, to view the 3D aligned structure
fragments in a PDB viewer.
in the red circle, to view the 3D aligned structure
fragments in a PDB viewer.
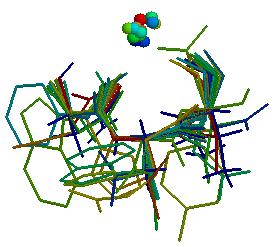
The picture shows a very good concentration of CA inside the loop.
The end