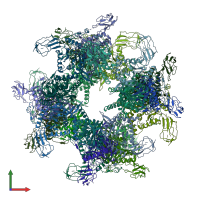
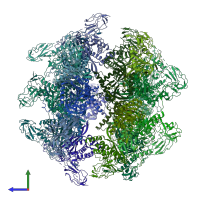
Function and Biology Details
Reaction catalysed:
Acetyl-CoA + enzyme N(6)-(dihydrolipoyl)lysine = CoA + enzyme N(6)-(S-acetyldihydrolipoyl)lysine
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
- Biotin/lipoyl attachment
- 2-oxo acid dehydrogenase, lipoyl-binding site
- Single hybrid motif
- Peripheral subunit-binding domain
- E3-binding domain superfamily
- 2-oxoacid dehydrogenase acyltransferase, catalytic domain
- Chloramphenicol acetyltransferase-like domain superfamily
- Dihydrolipoamide acetyltransferase pyruvate dehydrogenase complex
Structure analysis Details
Assembly composition:
homo 24-mer (preferred)
Assembly name:
PDBe Complex ID:
PDB-CPX-139312 (preferred)
Entry contents:
1 distinct polypeptide molecule
Macromolecule: