Carbo et al., (2013). Systems modeling of molecular mechanisms controlling cytokine-driven CD4+ T cell differentiation and phenotype plasticity.
December 2016, model of the month by Corina Dueñas Roca
Original model: BIOMD0000000451
Introduction
Quantitative modelling of molecular networks help us to define patterns of the components in a determined context. Therefore, a well characterized system could help us to identify potential therapeutic targets. The case presented here by Carbo et al.,(2013)[1] illustrates where mathematical modelling seeks to extend the current knowledge on CD4+ T-cell differentiation and plasticity with special focus in the switch from Th17 differentiations to iTreg. Keeping an equilibrium between cytokines involved in the immune response determines the condition of homoeostasis or disease.
In silico simulations were performed in this study, to predict the conditions of CD4+ T-cell differentiation. This model has been validated by in vitro and in vivo experiments. Finally, the key regulatory role of the nuclear protein PPARγ has been described in a dose-dependent manner. This protein determines the switch from Th17 phenotype to iTreg phenotype, and is a promising therapeutic target.
Model
The model aim to describe the dynamics of the CD4+ T-cell and the plasticity to four different phenotypes (Th1, Th2, Th17, and iTreg) (Figure 1). It is represented in an ordinary differential equation (ODE)-based computational model. Furthermore with the Hill function the effect of a ligand binding a macromolecule through cooperative binding is quantified, whereas the mass action laws represent the dynamic equilibriums for elementary reactions.
Results
The mathematical model describe the four possible phenotypes for CD4+ T-cells, where Th17 differentiation from a naive CD4+ T-cell can be induced by the combination of external IL-6 and TGFβ. Therefore in the T-cell there are up-regulation of RORγt, IL-17 and STAT3. PPARγ is a key regulatory factor in the CD4+ T-cell plasticity after sensitivity analysis. Furthermore, in vitro analysis on isolated CD4+ T-cells from spleens of wild type or PPARγ knock-down mice supported this evidence. Pioglitazone, a PPARγ agonist, shows experimentally that PPARγ switch the CD4+ T-cell differentiation from Th17 to iTreg in a dose dependent manner, increasing the levels of Foxp3 in Th17 cells (Figure 2).
In vivo analysis in mouse which lack PPARγ shows an increase of IL-17 and RORγ in Th17 cells, and mice treated with Pioglitazone recovered weight and their disease activity scores dropped significantly. Moreover mice treated with Pioglitazone had lower lymphocytic infiltration and crypt hyperplasia and showed a switch from a predominant Th17 into an iTreg phenotype. On the other hand untreated mice maintained a predominant Th17 response characterized by increased levels of CD4+ T-cells expressing RORγt and IL-17A (Figure 3). This data supports the in silico prediction that activation of PPARγ in Th17 cells favours differentiation into iTreg cells, which facilitates colonic tissue reconstitution and recovery from disease.

Figure 2. In silico activation of PPARγ in a T helper (Th)17 cell on the levels of FOXP3, IL-17 and RORγt.
Increasing concentrations of PPARγ in Th17 cells led to down-regulation of RORγt and IL17 and upregulation of FOXP3. Figure taken from [1].
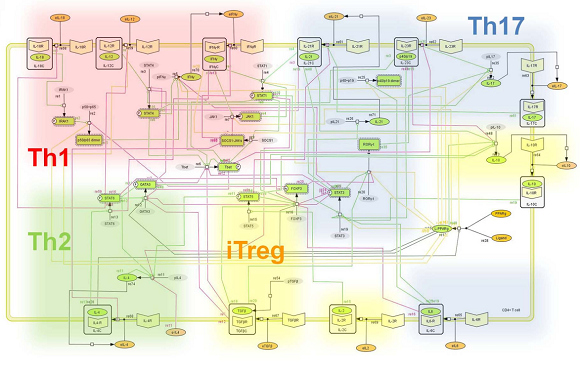
Figure 1. Network model illustrating the complex intracellular signaling pathways and transcriptional factors controlling the CD4+ T-cell differentiation process.
Ordinary differential equation (ODE)-based computational model. Representation of the network topologies associated with differentiation towards T helper Th1 (red cloud), Th2 (green cloud), Th17 (blue cloud) and iTreg1 (yellow cloud). Figure taken from [1].

Figure 3. Computational simulation of the effect of PPARγ deficiency on differentiation from a naive state into either Th17 or iTreg phenotypes
The loss of PPARγ in silico caused up-regulation of RORγt and IL-17 in Th17 (Figure 3B), and down-regulation of FOXP3 in iTreg cells (Figure 3D) compared to wild type CD4+ T-cells (Figure 3A and Figure 3C). Figure taken from [1]
The authors suggest that this model can be improved by adding stochastic analysis. This modelling approach allow us to narrow the design of experiments and to better understand the molecular mechanisms of action controlling CD4+ T-cell differentiation. Sensitivity analysis indicate that PPARγ is a promising therapeutic target for chronic inflammatory and infectious diseases where Th17 cells contribute to the gut immunopathogenesis.
Conclusion
The key role of PPARγ in regulating the Th-17 differentiation process predicted by the mathematical model presented here fits well with the already existing reports [2, 3, 4]. Surprisingly, these results fits better with in vivo results in mice with induced colitis rather than in vitro experiments. The explanation could be that the in vitro system lack other mechanisms that participate in the immune response.
References
- Carbo et al. Systems Modeling of Molecular Mechanisms Controlling Cytokine-Driven CD4+ T Cell Differentiation and Phenotype Plasticity. PLoS Comput Biol. 2013 April; 9(4): e1003027.
- Wohlfert et al. Peroxisome proliferator-activated receptor gamma (PPARgamma) and immunoregulation: enhancement of regulatory T cells through PPARgamma-dependent and -independent mechanisms. J Immunol. 2007 April; 178(7): 4129-35.
- Lei et al. Peroxisome proliferator-activated receptor α and γ agonists together with TGF-β convert human CD4+CD25- T cells into functional Foxp3+ regulatory T cells. J Immunol. 2010 December; 185(12): 7186-98.
- Klotz et al. The nuclear receptor PPAR gamma selectively inhibits Th17 differentiation in a T cell-intrinsic fashion and suppresses CNS autoimmunity. J Exp Med. 2009 September; 206(10): 2079-89.
