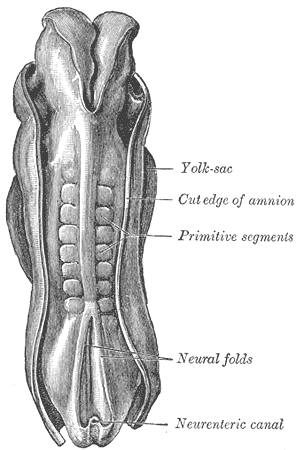
 Figure 1: Somites in a human embryo, dorsal view. From [2] | The vertebrate body is organised into segments, repetitive units along the anterior-posterior axis (as clearly seen, for instance, in the vertebral column). In embryonic development, the process of segmentation starts when mesodermal tissue alongside the neural tube assembles into repetitive blocks of cells. These blocks of cells are called somites and will later give rise to vertebrae, ribs, the dermis of the dorsal skin and the skeletal muscles of the back, body wall and limbs [1]. Figure 1 shows somites in a developing human embryo. Somitogenesis proceeds from rostral to caudal, with a new pair of somites forming every 90 minutes (reviewed in [3]). A diagram of somite formation is displayed in figure 2. The process is thought to be governed by a "molecular clock", with levels of some proteins in the presynaptic mesoderm oscillating prior to somite formation (reviewed in [3]), a model first proposed by Cooke and Zeeman in 1976 [4] (reviewed in [5]). Signalling pathways that have been implicated in this "molecular clock" include the Notch pathway, the Wnt pathway and the Fibroblast Growth factor (FGF) pathway. All of these pathways contain negative feedback loops that would allow them to generate oscillations. | The model proposed by Goldbeter and Pourquié ([6], BIOMD0000000201) explores the oscillations within these three signalling modules and their dynamic interactions.  Figure 3: Feedback loop in the Notch signalling pathway. Figure taken from [7]. | The Notch pathway allows for direct communication between neighbouring cells: Both the Notch receptor and its ligand Delta-Serrate-Lag-2 (DSL) are transmembrane proteins; binding takes place between a ligand molecule on one cell and the extracellular part of the Notch receptor on an adjacent cell (reviewed in [5]). Upon activation, the Notch intracellular domain (NICD) is cleaved and translocates into the nucleus, where it acts as part of a transcription factor complex (reviewed in [5]). Among the downstream targets of this transcription factor complex is the glycosyltransferase Lunatic fringe (Lfng), which, in turn, modulates the ligand affinity (and hence, activity) of the Notch receptor (reviewed in [5]). The resulting feedback loop can produce oscillations in the Notch signalling pathway (reviewed in [5], [7]), which is depicted in figure 3. | | The Wnt signalling pathway is triggered by binding of Wnt to the transmembrane receptor Frizzled and subsequent activation of Dishevelled (Dsh) (reviewed in [6]). Dsh stabilises β-catenin, because it prevents the formation of a "destruction complex" involving the kinase Gsk3, which would otherwise promote β-catenin degradation (reviewed in [6]). β-catenin can thus accumulate and translocate into the nucleus, where it promotes the expression of a number of target genes (reviewed in [6]). One of the targets of the Wnt/β-catenin cascade is Axin2, which has been shown to exhibit oscillations in the pre-somitic mesoderm [8]. Axin2 is a member of the β-catenin destruction complex, thus closing a negative feedback loop (reviewed in [6] and [7]), which is illustrated in figure 4. The oscillations in Axin2 levels arising from this negative feedback loop are out of phase with Lfng oscillations and are still maintained even if Notch signalling is disrupted [8]. |  Figure 4: Feedback loop in the Wnt signalling pathway. Figure modified from [6]. |  Figure 5: Feedback loop in the FGF signalling pathway. Figure modified from [6]. | The third oscillating pathway involved in somite segmentation is the Fibroblast Growth Factor (FGF) signalling pathway, triggered by FGF binding to the FGF receptor. This induces Ras activation, which further increases the activity of the MAP kinase ERK. Active ERK triggers the activation of a transcription factor, which induces the expression of phosphatase MKP3/Dusp6. This phosphatase inactivates ERK, thus providing a negative feedback loop (reviewed in [6]). The (simplified) FGF signalling pathway is illustrated in figure 5. In contrast to the Notch and Wnt feedback loops, the feedback loop in the FGF pathway does not act on protein levels, but on the prevalence of phosphorylation [6]. | The model by Goldbeter and Pourquié [6] illustrates how, while all three modules can generate oscillations independently of each other, their coupling through common intermediates can, under some conditions, produce synchronised oscillations. Figures 6 and 7 illustrates the difference between uncoupled and coupled oscillations of all three modules. The model combines three well-described pathways and provides a basis for closer examination of their interactions and interregulation. |  Figure 2: Somite formation diagram. From [3]
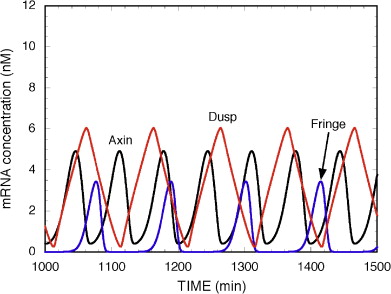 Figure 6: Oscillations in the Notch, Wnt and FGF pathways without coupling. From [6].
 Figure 7: Coupled oscillations in the Notch, Wnt and FGF signalling pathways. From [6]. |







