Urease
Ureases hydrolyse urea into ammonia and carbamate. Ureases have been isolated from a wide range of bacteria, fungi and higher plants where it allows the organism to use urea as a nitrogen source. Ureases uses an almost unique bi-nickel catalytic centre which is liganded by a carbamylated lysine.
The mechanism of this enzyme has been subject to debate since the early 1920s and the precise steps in catalysis remain unclear [PMID:20471401].
Reference Protein and Structure
- Sequences
-
P18316
 (3.5.1.5)
(3.5.1.5)
P18315 (3.5.1.5)
(3.5.1.5)
P18314 (3.5.1.5)
(3.5.1.5)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
[Enterobacter] aerogenes (Bacteria)

- PDB
-
1fwj
- KLEBSIELLA AEROGENES UREASE, NATIVE
(2.2 Å)



- Catalytic CATH Domains
-
3.20.20.140
 (see all for 1fwj)
(see all for 1fwj)
- Cofactors
- Nickel(2+) (2), Water (3) Metal MACiE
Enzyme Mechanism
Introduction
The active site contains two nickels. Carbamylated lysine 217 provides an oxygen ligand to each nickel. One nickel ion is coordinated by three ligands: two histidines (246 and 272) and the carbamylated lysine 217 (with low occupancy of a fourth ligand) and the second is coordinated by five ligands: two histidines (136 and 134), aspartate 360, a water and the carbamylated lysine 217. Urea enters the active site and ligates to Ni-1 to complete its tetrahedral coordination. It also forms a hydrogen bond to histidine 219 which polarises the urea carbonyl. Histidine 320 acts as a general base and abstracts a proton from the Ni-2 water ligand, the resulting hydroxide ligand of Ni-2 then attacks the urea carbonyl carbon. The resulting tetrahedral intermediate decomposes, with the assistance of an unidentified general acid, to ammonia and Ni bound carbamylate which then dissociates.
Catalytic Residues Roles
| UniProt | PDB* (1fwj) | ||
| Lys217 (ptm) | Kcx217C (ptm) | Post-translationally modified lysine residue. Acts as a bridging ligand between the two Ni(II) ions. | activator, metal ligand |
| Asp360 | Asp360C | Forms part of the nickel 1 binding site, also helps activate His320. | metal ligand |
| His134, His136 | His134C, His136C | Forms part of the nickel 1 binding site. | metal ligand |
| His246, His272 | His246C, His272C | Forms part of the nickel 2 binding site. | metal ligand |
| His320, His219 | His320C, His219C | Acts as a general acid/base. | proton acceptor, proton donor |
| Asp221 | Asp221C | Acts as a general acid/base, activating His219 . | activator, proton acceptor, proton donor |
| Arg336 | Arg336C | Helps stabilise the charge in the active site, activating both His320 and Asp221. | activator |
Chemical Components
proton transfer, bimolecular nucleophilic addition, deamination, unimolecular elimination by the conjugate base, reaction occurs outside the enzyme, intramolecular eliminationReferences
- Balasubramanian A et al. (2010), J Mol Biol, 400, 274-283. Crystal Structure of the First Plant Urease from Jack Bean: 83 Years of Journey from Its First Crystal to Molecular Structure. DOI:10.1016/j.jmb.2010.05.009. PMID:20471401.
- Roberts BP et al. (2012), J Am Chem Soc, 134, 9934-9937. Wide-Open Flaps Are Key to Urease Activity. DOI:10.1021/ja3043239. PMID:22670767.
- Carlsson H et al. (2010), Bioinorg Chem Appl, 2010, 1-8. Computational Modeling of the Mechanism of Urease. DOI:10.1155/2010/364891. PMID:20886006.
- Krajewska B (2009), J Mol Catal B Enzym, 59, 9-21. Ureases I. Functional, catalytic and kinetic properties: A review. DOI:10.1016/j.molcatb.2009.01.003.
- Barrios AM et al. (2000), J Am Chem Soc, 122, 9172-9177. Interaction of Urea with a Hydroxide-Bridged Dinuclear Nickel Center: An Alternative Model for the Mechanism of Urease. DOI:10.1021/ja000202v.
- Pearson MA et al. (2000), Biochemistry, 39, 8575-8584. Kinetic and Structural Characterization of Urease Active Site Variants†,‡. DOI:10.1021/bi000613o. PMID:10913264.
- Pearson MA et al. (1998), Biochemistry, 37, 6214-6220. Chemical Rescue ofKlebsiella aerogenesUrease Variants Lacking the Carbamylated-Lysine Nickel Ligand†,‡. DOI:10.1021/bi980021u. PMID:9558361.
- Park IS et al. (1996), J Biol Chem, 271, 18632-18637. Characterization of the Mononickel Metallocenter in H134A Mutant Urease. DOI:10.1074/jbc.271.31.18632. PMID:8702515.
- Jabri E et al. (1996), Biochemistry, 35, 10616-10626. Structures of theKlebsiella aerogenesUrease Apoenzyme and Two Active-Site Mutants†,‡. DOI:10.1021/bi960424z. PMID:8718850.
- Jabri E et al. (1995), Science, 268, 998-1004. The crystal structure of urease from Klebsiella aerogenes. DOI:10.1126/science.7754395. PMID:7754395.
- Park IS et al. (1993), Protein Sci, 2, 1034-1041. Site-directed mutagenesis ofKlebsiella aerogenesurease: Identification of histidine residues that appear to function in nickel ligation, substrate binding, and catalysis. DOI:10.1002/pro.5560020616. PMID:8318888.
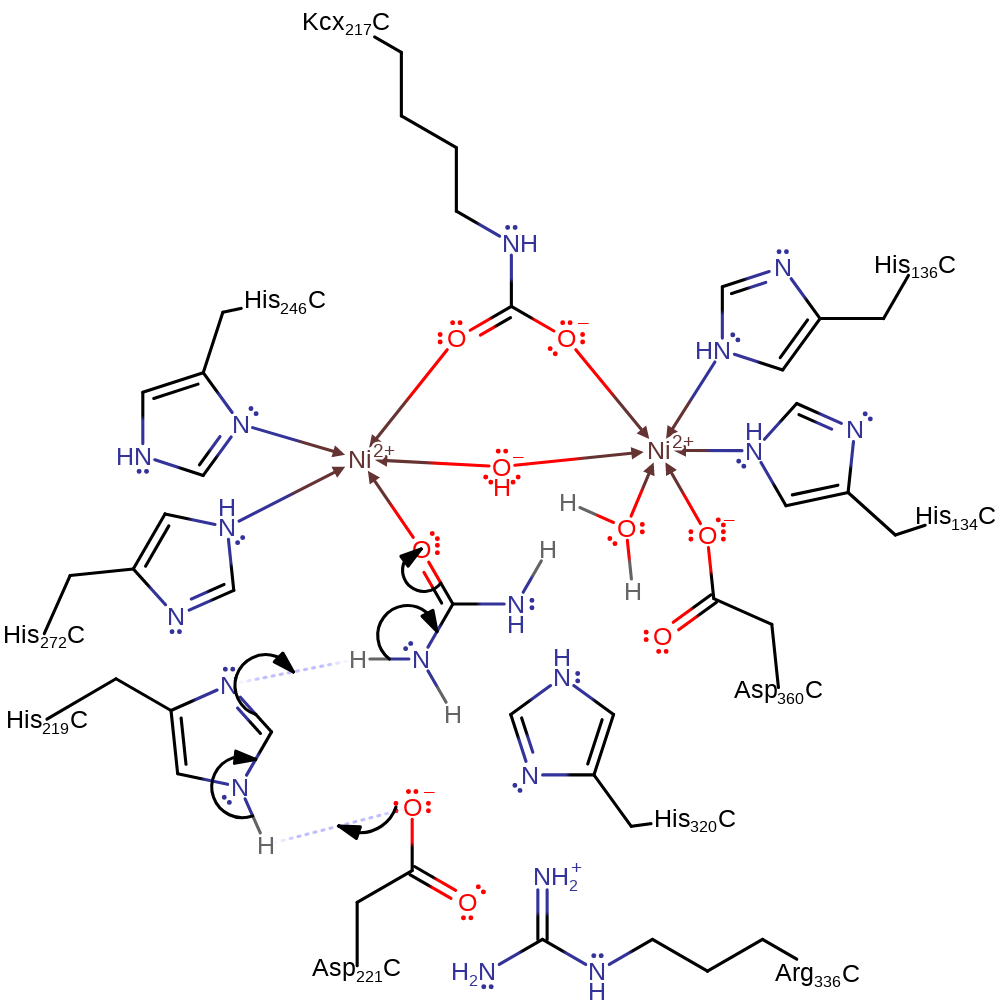
Step 1. Asp221 deprotonates His219, which in turn deprotonates the nickel bound urea, forming the active conformer for this mechanism.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| Kcx217C (ptm) | metal ligand |
| Kcx217C (ptm) | activator |
| Arg336C | activator |
| His219C | proton acceptor, proton donor |
| Asp221C | proton acceptor |
| His219C | proton relay |
Chemical Components
proton transfer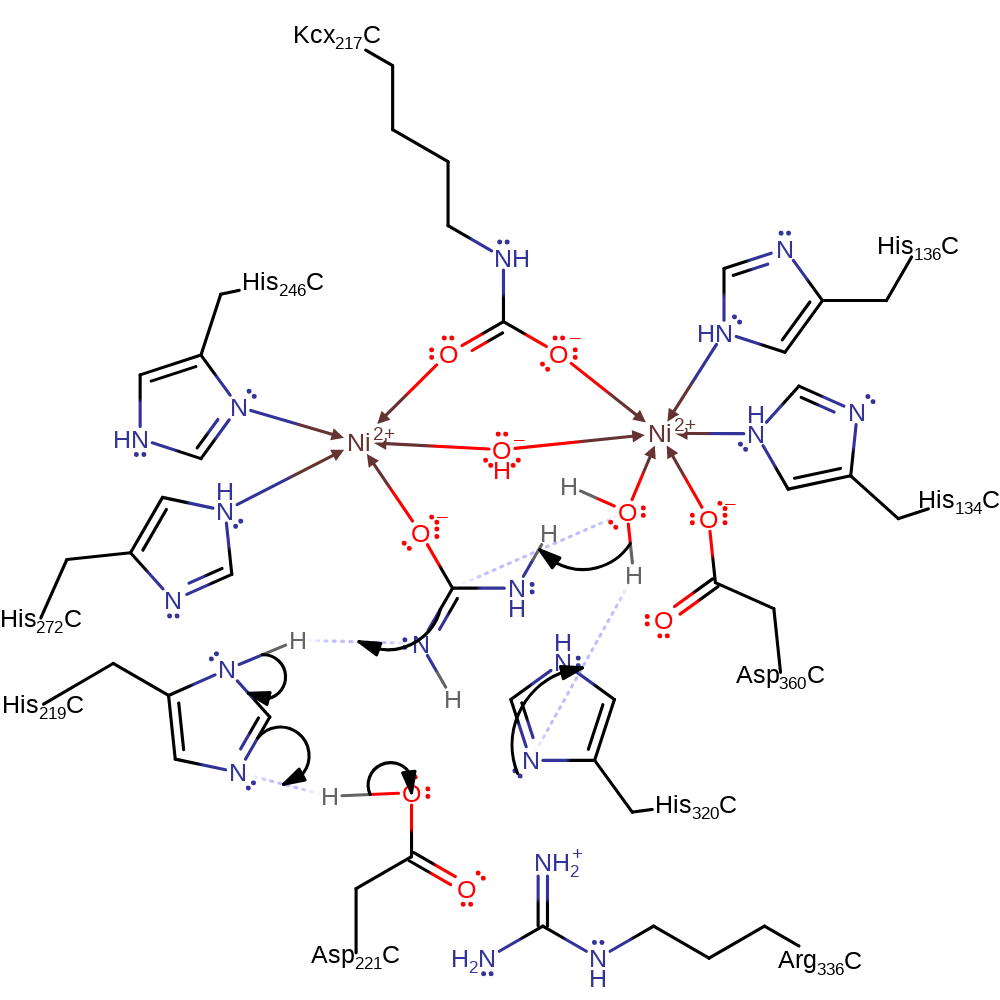
Step 2. His320 deprotonates nickel bound water, which initiates a nucleophilic attack on the nickel bound urea in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Kcx217C (ptm) | activator, metal ligand |
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| His219C | proton acceptor, proton donor |
| His320C | proton acceptor |
| Asp221C | proton donor |
| His219C | proton relay |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer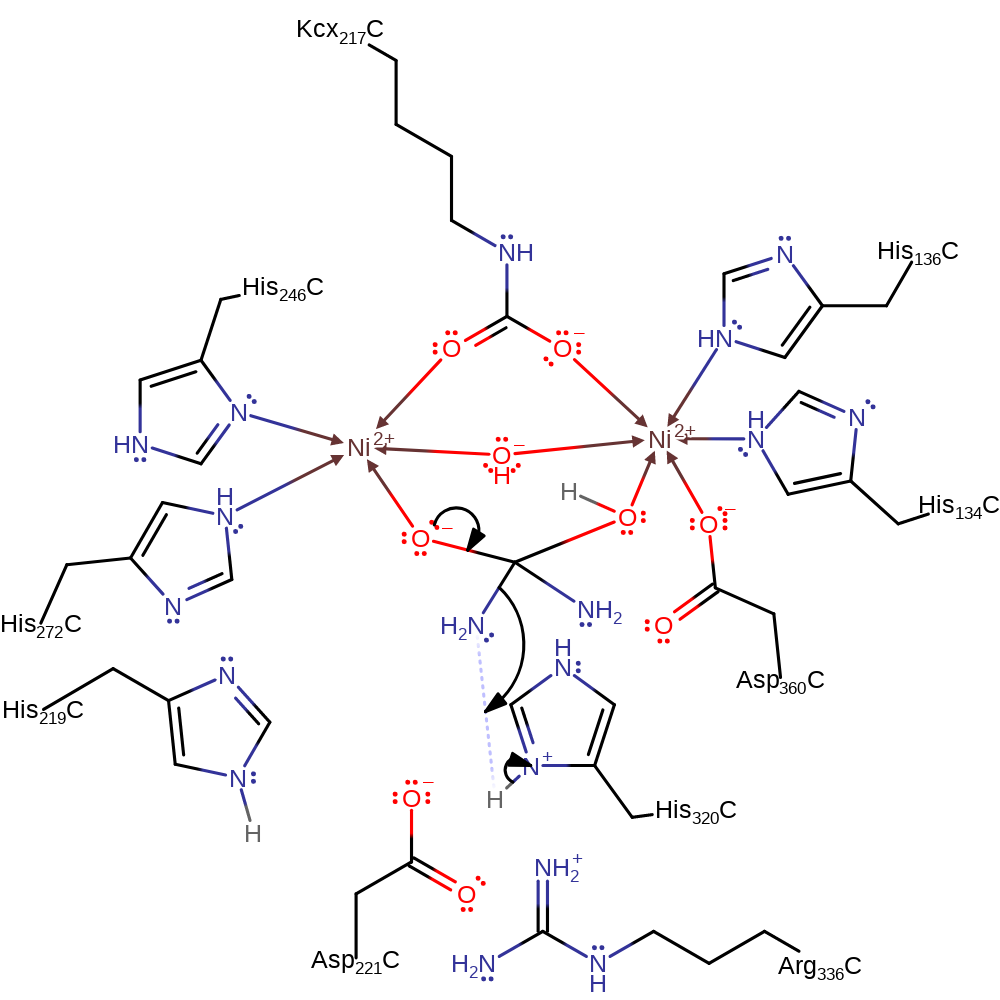
Step 3. The tetrahedral intermediate collapses, eliminating ammonia with concomitant deprotonation of His320.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| Kcx217C (ptm) | metal ligand |
| Asp221C | activator |
| Arg336C | activator |
| Kcx217C (ptm) | activator |
| His320C | proton donor |
Chemical Components
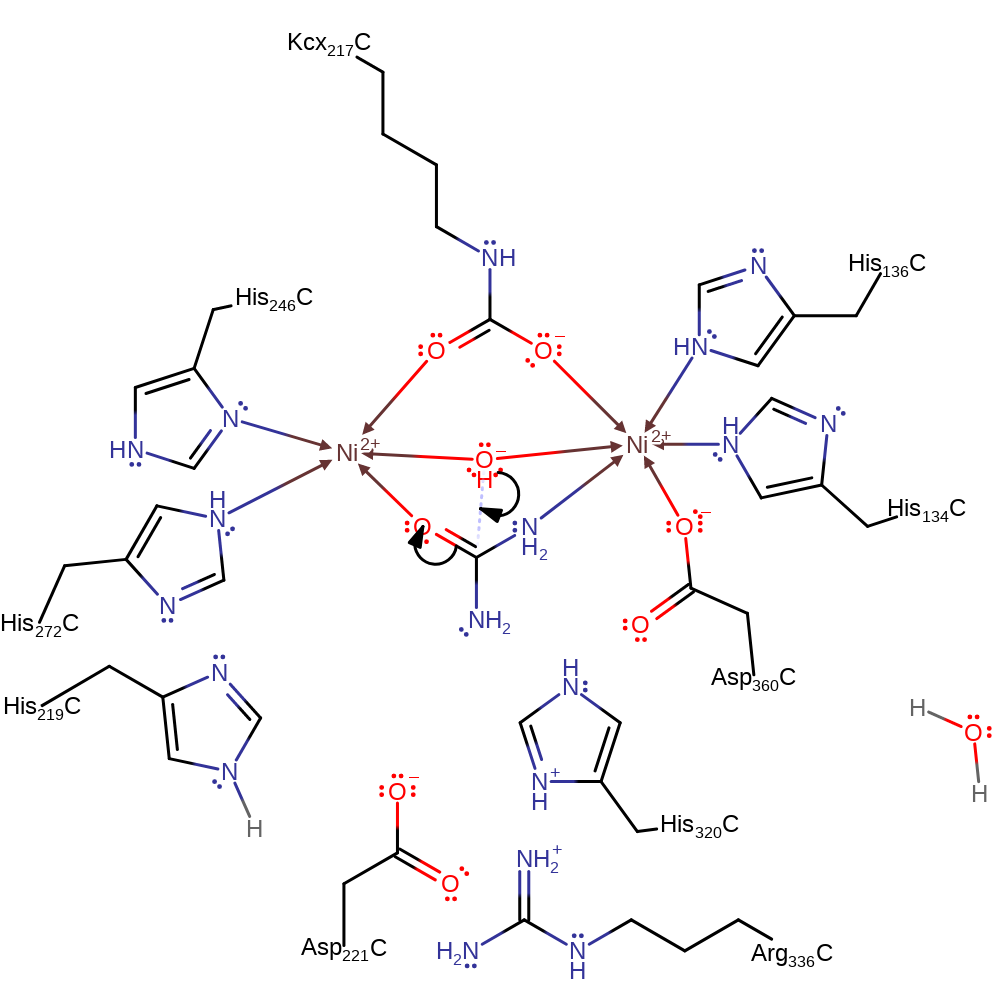
deamination, ingold: unimolecular elimination by the conjugate base
Step 4. Carbamate dissociates from the active site and is spontaneously converted to carbon dioxide and a second molecule of ammonia
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, ingold: intramolecular eliminationIntroduction
A third proposal is the elimination reaction from Barios and Lippard [PMID:11300826] has also been proposed on the observation of cyanic intermediates. Theoretical work by Estiu and Merz [PMID:16584179, PMID:17676790] suggests that the elimination pathway may occur in competition with the more traditionally proposed mechanisms.
Catalytic Residues Roles
| UniProt | PDB* (1fwj) | ||
| Lys217 (ptm) | Kcx217C (ptm) | Post-translationally modified lysine residue. Acts as a bridging ligand between the two Ni(II) ions. | metal ligand |
| Asp360 | Asp360C | Forms part of the nickel 1 binding site, also helps activate His320. | activator, metal ligand |
| His134, His136 | His134C, His136C | Forms part of the nickel 1 binding site. | metal ligand |
| His246, His272 | His246C, His272C | Forms part of the nickel 2 binding site. | metal ligand |
| His320, His219 | His320C, His219C | Acts as a general acid/base. | proton acceptor, proton donor |
| Asp221 | Asp221C | Acts as a general acid/base, deprotonating His219 to activate it. It later donates this proton to His320. | proton acceptor, proton donor |
| Arg336 | Arg336C | Helps stabilise the charge in the active site, activating both His320 and Asp221. | activator |
Chemical Components
reaction occurs outside the enzyme, bimolecular nucleophilic addition, inferred reaction step, intramolecular elimination, native state of enzyme regeneratedReferences
- Barrios AM et al. (2001), Inorg Chem, 40, 1250-1255. Decomposition of Alkyl-Substituted Urea Molecules at a Hydroxide-Bridged Dinickel Center. DOI:10.1021/ic000933w. PMID:11300826.
- Roberts BP et al. (2012), J Am Chem Soc, 134, 9934-9937. Wide-Open Flaps Are Key to Urease Activity. DOI:10.1021/ja3043239. PMID:22670767.
- Krajewska B (2009), J Mol Catal B Enzym, 59, 9-21. Ureases I. Functional, catalytic and kinetic properties: A review. DOI:10.1016/j.molcatb.2009.01.003.
- Estiu G et al. (2007), J Phys Chem B, 111, 10263-10274. Competitive Hydrolytic and Elimination Mechanisms in the Urease Catalyzed Decomposition of Urea. DOI:10.1021/jp072323o. PMID:17676790.
- Estiu G et al. (2006), Biochemistry, 45, 4429-4443. Catalyzed Decomposition of Urea. Molecular Dynamics Simulations of the Binding of Urea to Urease†. DOI:10.1021/bi052020p. PMID:16584179.
- Park IS et al. (1996), J Biol Chem, 271, 18632-18637. Characterization of the Mononickel Metallocenter in H134A Mutant Urease. DOI:10.1074/jbc.271.31.18632. PMID:8702515.

Step 1. Asp221 deprotonates His219, which in turn abstracts a proton from the Ni(II) bound urea, forming a negatively charged intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| Arg336C | activator |
| Kcx217C (ptm) | metal ligand |
| Asp221C | proton acceptor |
| His219C | proton acceptor, proton donor, proton relay |
Chemical Components
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| Arg336C | activator |
| Asp360C | activator |
| Kcx217C (ptm) | metal ligand |
| Asp221C | proton donor |
| His320C | proton acceptor |
Chemical Components

Step 3. The negatively charged intermediate collapses, eliminating ammoinia, which abstracts the proton from His320.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| Arg336C | activator |
| Asp360C | activator |
| Kcx217C (ptm) | metal ligand |
| His320C | proton donor |
Chemical Components

Step 4. Cyanic acid is the product of this enzyme mechanism, thus it is likely that the final hydrolysis occurs outside of the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, ingold: bimolecular nucleophilic addition, inferred reaction step
Step 5. Ammonia is eliminated in the final non-enzyme catalysed step of the reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, ingold: intramolecular elimination, inferred reaction step
Step 6. Inferred step to regenerate the enzyme's active site. Two water molecules displace the product to form the ground state. It is unclear how the His219 returns to it's native state, we have represented a water facilitated tautomerisation reaction here.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| Kcx217C (ptm) | metal ligand |
| His219C | proton relay, proton acceptor, proton donor |
Chemical Components
inferred reaction step, native state of enzyme regeneratedIntroduction
Proposed by Benini et al. for Bacillus pasteurii enzyme, urea binds in a bidentate manner with its carbonyl oxygen bound to Ni1 and one of the amino group bound to Ni2, thus replacing three water moieties, leaving only the bridging hydroxide. This hydroxide attacks urea to give the tetrahedral transition state leading to formation of ammonia and carbamate.
Catalytic Residues Roles
| UniProt | PDB* (1fwj) | ||
| Lys217 (ptm) | Kcx217C (ptm) | Post-translationally modified lysine residue. Acts as a bridging ligand between the two Ni(II) ions. | activator, metal ligand |
| Asp360 | Asp360C | Forms part of the nickel 1 binding site, also helps activate His320. | metal ligand |
| His320 | His320C | Acts as a general acid/base. | proton acceptor, proton donor |
| His134, His136 | His134C, His136C | Forms part of the nickel 1 binding site. | metal ligand |
| His246, His272 | His246C, His272C | Forms part of the nickel 2 binding site. | metal ligand |
| His219 | His219C | Has no assigned role in this mechanism. | |
| Asp221, Arg336 | Asp221C, Arg336C | Activates His320. | activator |
Chemical Components
proton transfer, unimolecular elimination by the conjugate base, inferred reaction step, reaction occurs outside the enzyme, intramolecular eliminationReferences
- Benini S et al. (2001), J Biol Inorg Chem, 6, 778-790. Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism. DOI:10.1007/s007750100254. PMID:11713685.
- Roberts BP et al. (2012), J Am Chem Soc, 134, 9934-9937. Wide-Open Flaps Are Key to Urease Activity. DOI:10.1021/ja3043239. PMID:22670767.
- Carlsson H et al. (2010), Bioinorg Chem Appl, 2010, 1-8. Computational Modeling of the Mechanism of Urease. DOI:10.1155/2010/364891. PMID:20886006.
- Krajewska B (2009), J Mol Catal B Enzym, 59, 9-21. Ureases I. Functional, catalytic and kinetic properties: A review. DOI:10.1016/j.molcatb.2009.01.003.
- Benini S et al. (2004), J Am Chem Soc, 126, 3714-3715. Molecular Details of Urease Inhibition by Boric Acid: Insights into the Catalytic Mechanism. DOI:10.1021/ja049618p. PMID:15038715.
- Benini S et al. (1999), Structure, 7, 205-216. A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels. DOI:10.1016/s0969-2126(99)80026-4. PMID:10368287.
- Pearson MA et al. (1997), Biochemistry, 36, 8164-8172. Structures of Cys319 Variants and Acetohydroxamate-InhibitedKlebsiella aerogenesUrease†,‡. DOI:10.1021/bi970514j. PMID:9201965.
- Park IS et al. (1996), J Biol Chem, 271, 18632-18637. Characterization of the Mononickel Metallocenter in H134A Mutant Urease. DOI:10.1074/jbc.271.31.18632. PMID:8702515.
Step 1. Urea binds in a bindentate manner, displacing the active site water molecules. The bridging hydroxide group initiates a nucleophillic attack on the carbamyl carbon, forming a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg336C | activator |
| Kcx217C (ptm) | activator, metal ligand |
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
Chemical Components
proton transfer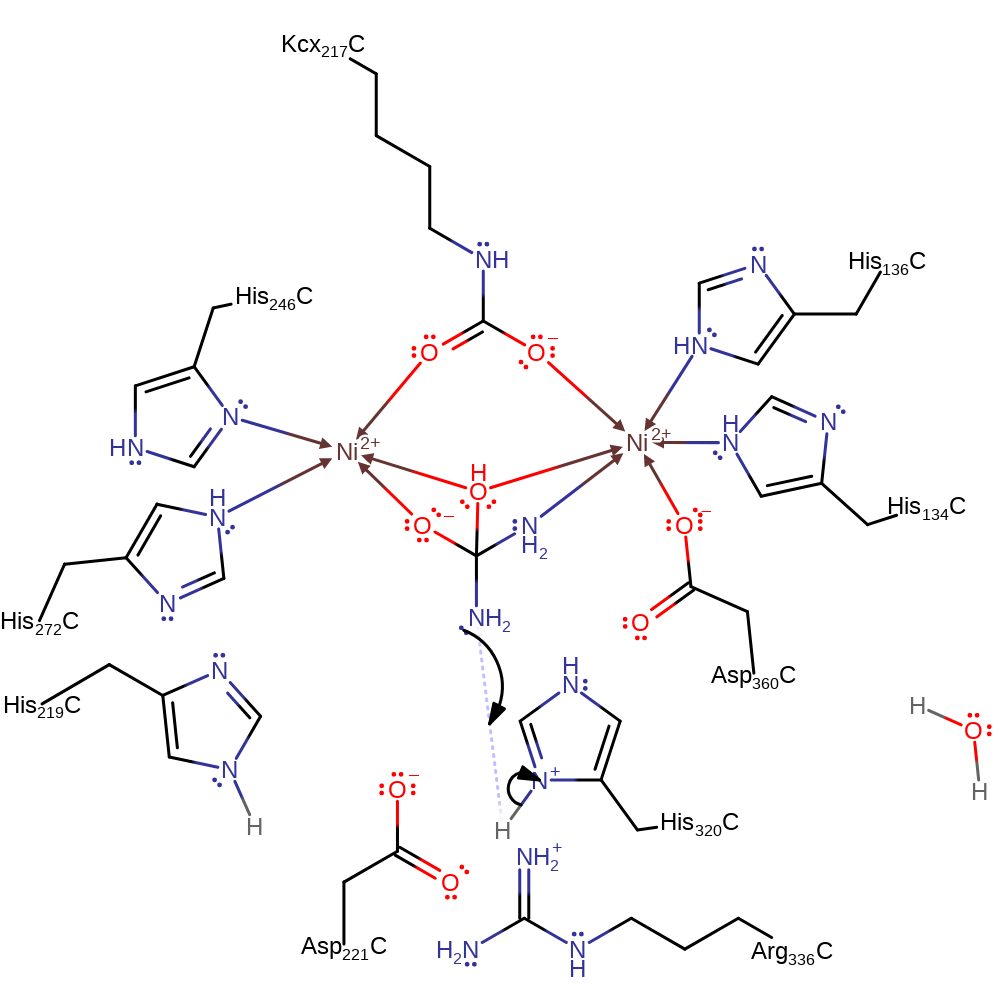
Step 2. The unbound NH2 group of the tetrahedral intermediate deprotonates His320.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| Kcx217C (ptm) | activator, metal ligand |
| Asp221C | activator |
| Arg336C | activator |
| His320C | proton donor |
Chemical Components
proton transfer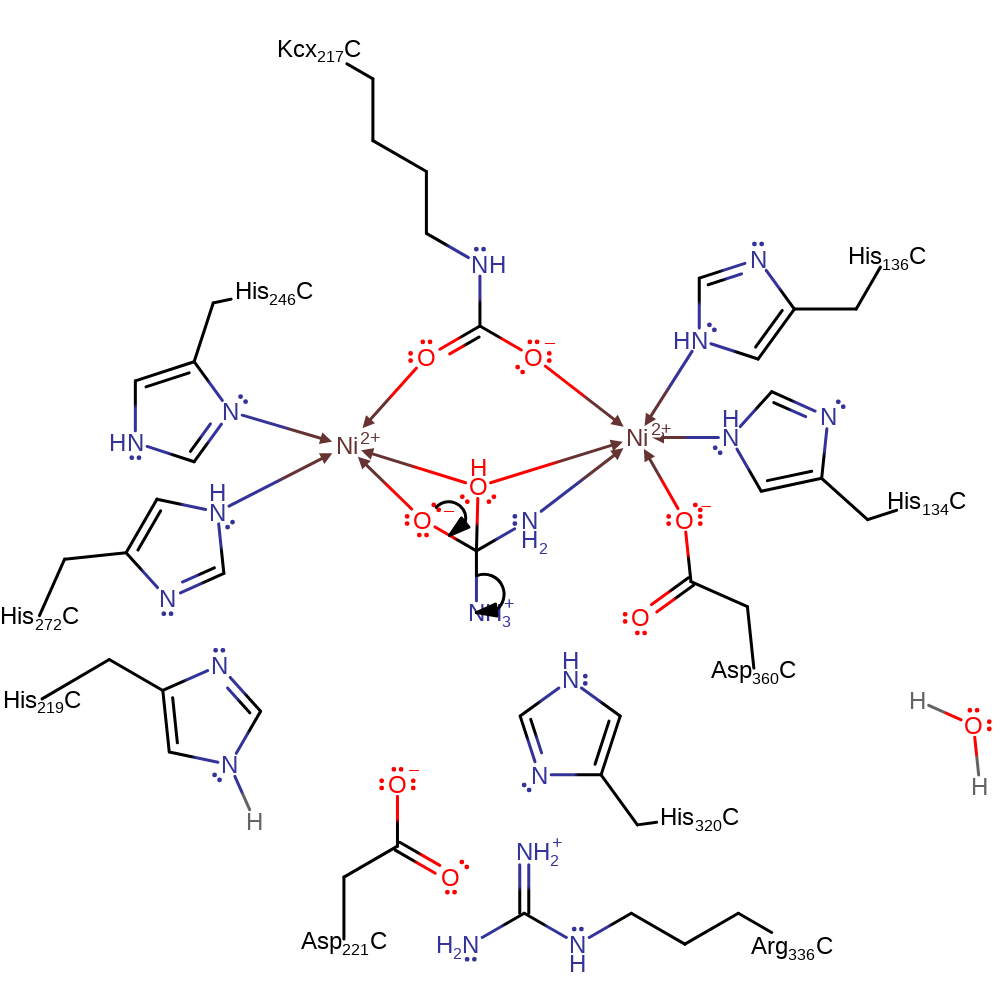
Step 3. The tetrahedral intermediate collapses, eliminating ammonia. The products then dissociate from the active site, replaced by water molecules.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| Kcx217C (ptm) | activator, metal ligand |
Chemical Components
ingold: unimolecular elimination by the conjugate base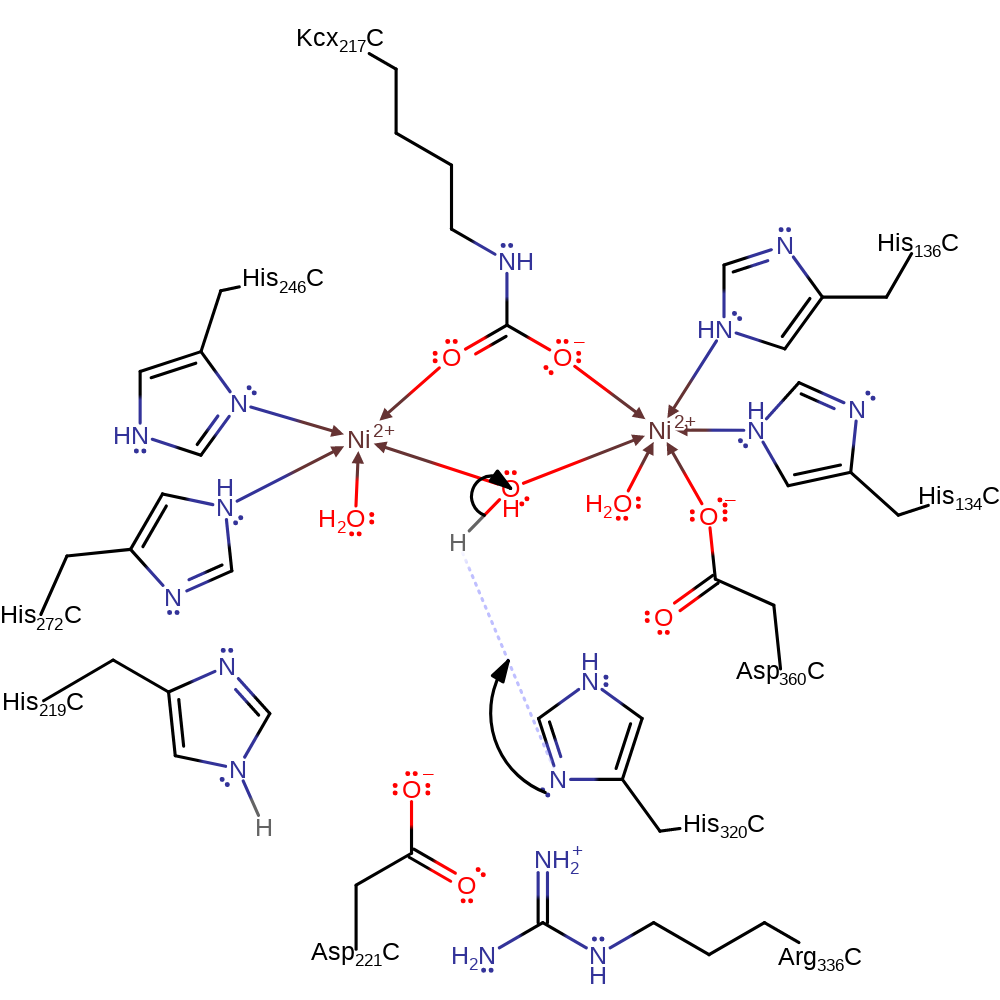
Step 4. In an inferred step, His320 abstracts a proton from one of the incoming water molecules to regenerate the enzyme's active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His134C | metal ligand |
| His136C | metal ligand |
| Asp360C | metal ligand |
| His246C | metal ligand |
| His272C | metal ligand |
| Kcx217C (ptm) | metal ligand |
| Asp221C | activator |
| His320C | proton acceptor |
Chemical Components
inferred reaction step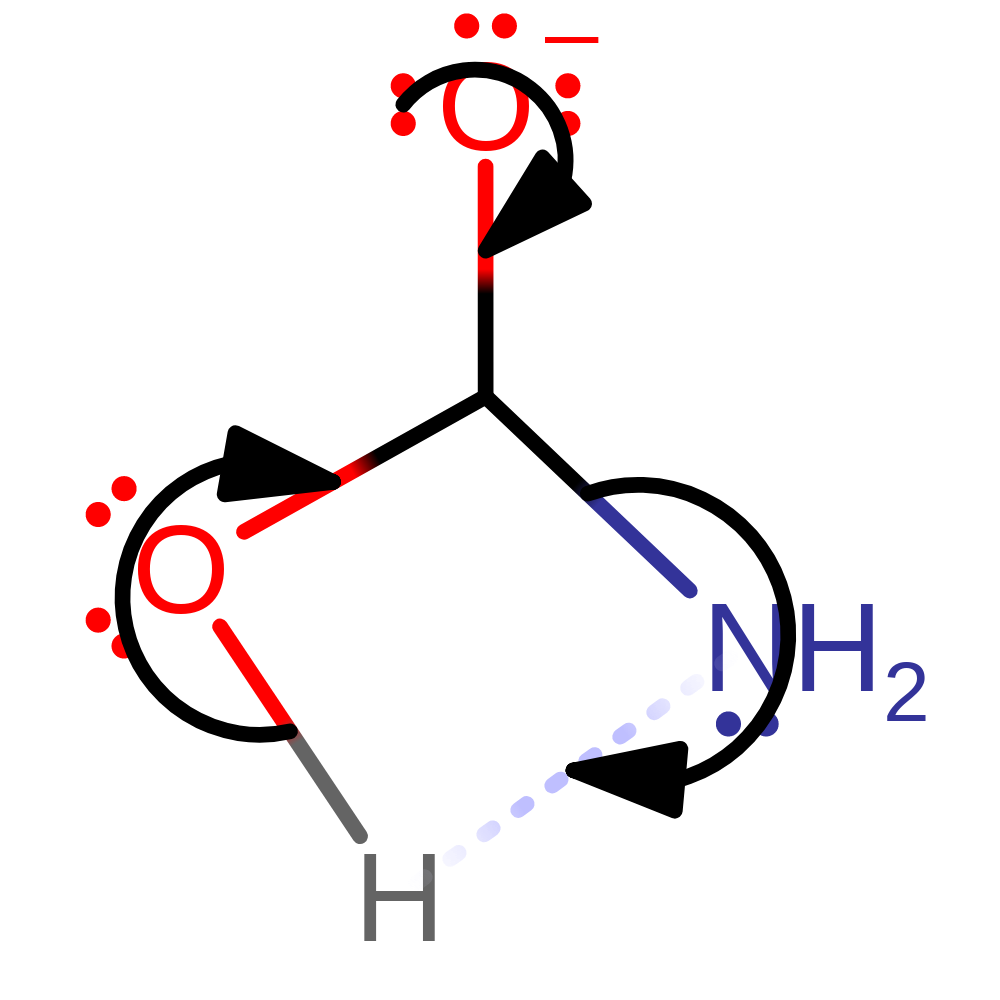
Step 5. The carbamate product rearranges outside of the active site to produce carbon dioxide and ammonia.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|



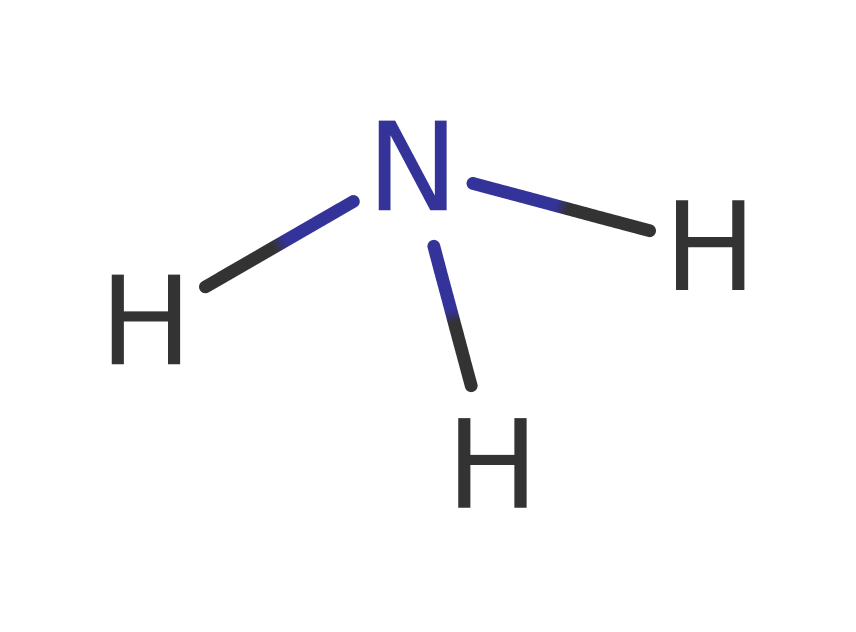
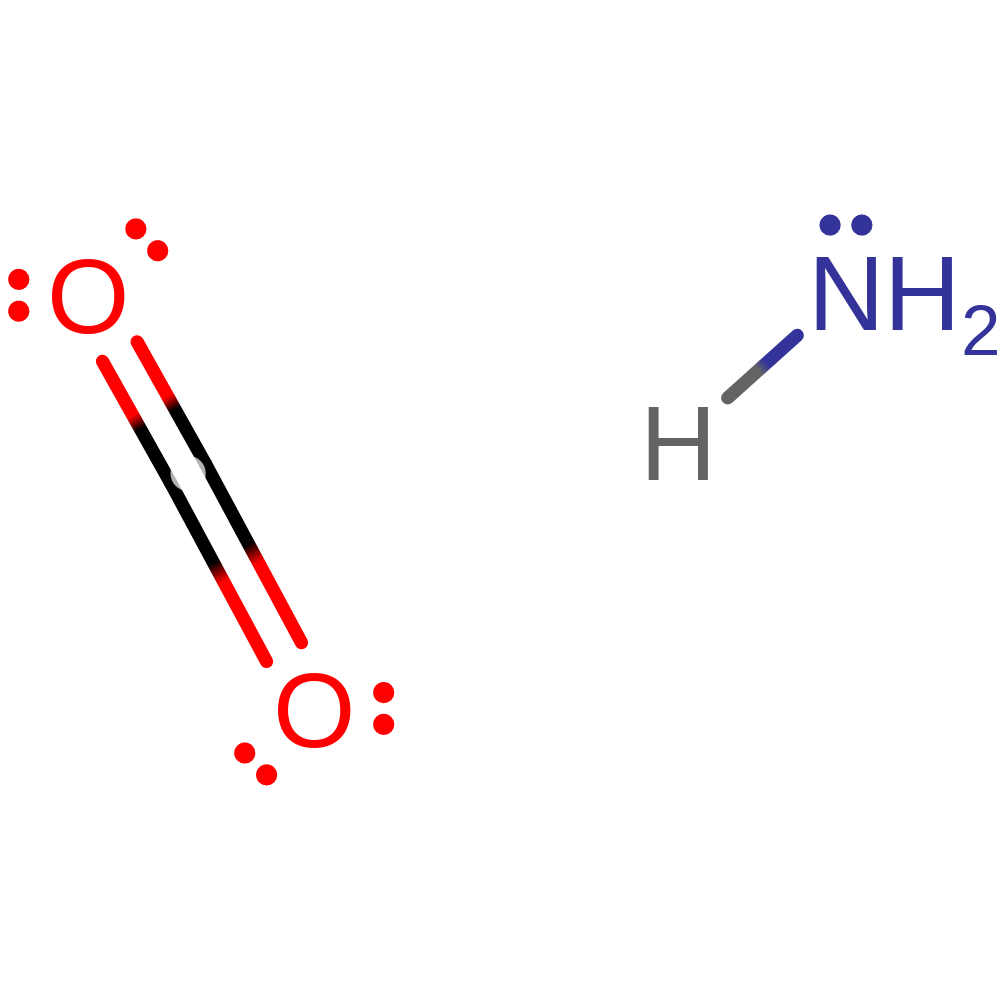 Download:
Download: 
 Download:
Download: 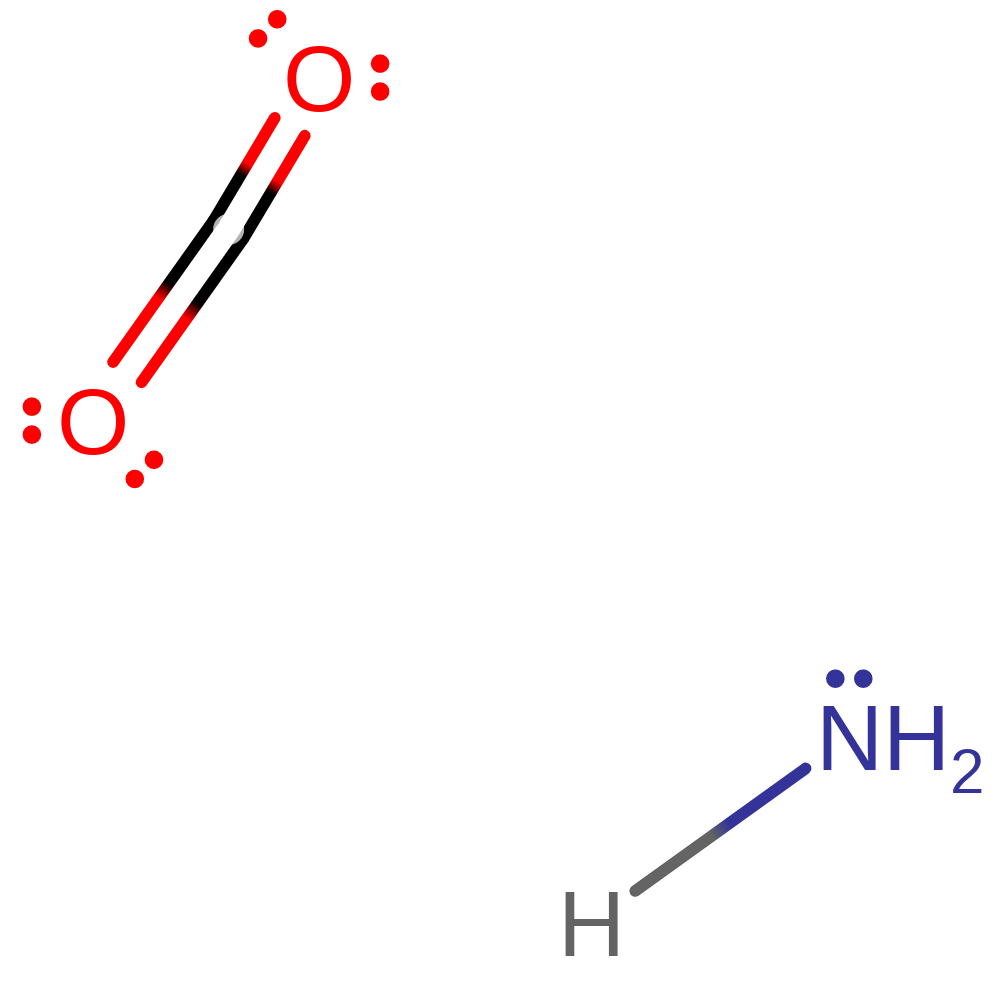 Download:
Download: