Uridine nucleosidase (family I)
Uracil-DNA glycosylase (UDG) are monofunctional glycosylases and initiate the base excision repair (BER) pathway for uracil by hydrolysing the N-C'1 glycosylic bond between a target uracil and an abasic site. The human BER cycle is important for restoring the chemical integrity of DNA.
The uracil-DNA glycosylase superfamily consists of 6 smaller families, based on sequence alignments. Both human and E.coli UDG come under Family I and are also called UNGs.
The enzyme mechanism has been hotly debated. Classically, an acid/base mechanism has been employed (mechanism proposal 2), but new evidence suggests a steric distortion of the DNA substrate catalyses the reaction in a mechanism similar to SN1 dissociation (mechanism proposal 1).
Reference Protein and Structure
- Sequence
-
P12295
 (3.2.2.27)
(3.2.2.27)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1eug
- CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED
(1.6 Å)



- Catalytic CATH Domains
-
3.40.470.10
 (see all for 1eug)
(see all for 1eug)
Enzyme Reaction (EC:3.2.2.3)
Enzyme Mechanism
Introduction
This mechanism represents the steric contortion mechanism. Here the tetrahedral distortion is imposed by the structurally rigid walls of the active site, formed by Tyr and Phe residues. The enzyme centre flattens the pucker ring of the uridine deoxyribose, raising the glycosylic bond to a semi-axial position, allowing pi-sigma* overlap. This stereoelectronic effect increases in strength as the substrate is further distorted towards the transition state. As the transition state move towards a tetrahedral geometry, and glycosylic bond rotation occurs, an anomeric effect is coupled to the pi systems of the uracil ring, resulting in orbital overlap of the glycosylic bond and the carbonyl C2 and C4 pi systems. The developing negative charge of the transition state is stabilised by hydrogen bonding to His-187. The enzyme funnels binding energy for use is catalysis by employing substrate distortions to couple two stereoelectronic effects to promote efficient catalysis. The abasic nucleotide relaxes into a more puckered conformation and withdraws from the enzyme while uracil tilts deeper into the active site, improving its stacking interactions with the Phe residue present. These rearrangements lower the product's energies relative to the reactant's, reducing the strain within the active site and allowing the enzyme to bind preferentially to the products.
Catalytic Residues Roles
| UniProt | PDB* (1eug) | ||
| Tyr66 | Tyr66A | The residue acts to distort the DNA substrate towards the transition state conformation through steric interactions with the pucker ring of the uridine deoxyribose. The residue is also implicated in preventing the binding of thymine within the active site. | activator, steric role |
| Phe77 | Phe77A | The residue acts to distort the DNA substrate towards the transition state conformation by steric and pi stacking interactions with the pucker ring of the uridine deoxyribose. | activator, steric role |
| His187 | His187A | The residue hydrogen bonds to the developing anion within the transition state, lowering its energy. | hydrogen bond donor, electrostatic stabiliser |
| Asp64 | Asp64A | Stabilising and activating the catalytic histidine. May also act as a general acid/base | activator, electrostatic stabiliser |
Chemical Components
intramolecular elimination, intermediate formation, overall reactant used, bimolecular nucleophilic addition, proton transfer, overall product formed, native state of enzyme regenerated, intermediate terminated, tautomerisation (not keto-enol), reaction occurs outside the enzymeReferences
- Parikh SS et al. (2000), Proc Natl Acad Sci U S A, 97, 5083-5088. Uracil-DNA glycosylase-DNA substrate and product structures: Conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. DOI:10.1073/pnas.97.10.5083. PMID:10805771.
- Schormann N et al. (2014), Protein Sci, 23, 1667-1685. Uracil-DNA glycosylases-Structural and functional perspectives on an essential family of DNA repair enzymes. DOI:10.1002/pro.2554. PMID:25252105.
- Drohat AC et al. (2014), Org Biomol Chem, 12, 8367-8378. Mechanisms for enzymatic cleavage of the N-glycosidic bond in DNA. DOI:10.1039/c4ob01063a. PMID:25181003.
- Jiang YL et al. (2003), Biochemistry, 42, 1922-1929. Powering DNA Repair through Substrate Electrostatic Interactions†. DOI:10.1021/bi027014x. PMID:12590578.
- Jiang YL et al. (2002), J Biol Chem, 277, 15385-15392. Probing the Limits of Electrostatic Catalysis by Uracil DNA Glycosylase Using Transition State Mimicry and Mutagenesis. DOI:10.1074/jbc.m200634200. PMID:11859082.
- Handa P et al. (2002), Nucleic Acids Res, 30, 3086-3095. Effects of mutations at tyrosine 66 and asparagine 123 in the active site pocket of Escherichia coli uracil DNA glycosylase on uracil excision from synthetic DNA oligomers: evidence for the occurrence of long-range interactions between the enzyme and substrate. PMID:12136091.
- Stivers JT et al. (2001), Arch Biochem Biophys, 396, 1-9. Uracil DNA Glycosylase: Insights from a Master Catalyst. DOI:10.1006/abbi.2001.2605. PMID:11716455.
- Jiang YL et al. (2001), Biochemistry, 40, 7710-7719. Reconstructing the substrate for uracil DNA glycosylase: tracking the transmission of binding energy in catalysis. PMID:11412125.
- Dinner AR et al. (2001), Nature, 413, 752-755. Uracil-DNA glycosylase acts by substrate autocatalysis. DOI:10.1038/35099587. PMID:11607036.
- Parikh SS et al. (2000), Mutat Res, 460, 183-199. Lessons learned from structural results on uracil-DNA glycosylase. DOI:10.1016/s0921-8777(00)00026-4. PMID:10946228.
- Werner RM et al. (2000), Biochemistry, 39, 14054-14064. Kinetic isotope effect studies of the reaction catalyzed by uracil DNA glycosylase: evidence for an oxocarbenium ion-uracil anion intermediate. PMID:11087352.
- Xiao G et al. (1999), Proteins, 35, 13-24. Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited. DOI:10.2210/pdb1eug/pdb. PMID:10090282.
- Kavli B et al. (1996), EMBO J, 15, 3442-3447. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. PMID:8670846.
- Mol CD et al. (1995), Cell, 80, 869-878. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. PMID:7697717.
- Savva R et al. (1995), Nature, 373, 487-493. The structural basis of specific base-excision repair by uracil–DNA glycosylase. DOI:10.1038/373487a0. PMID:7845459.
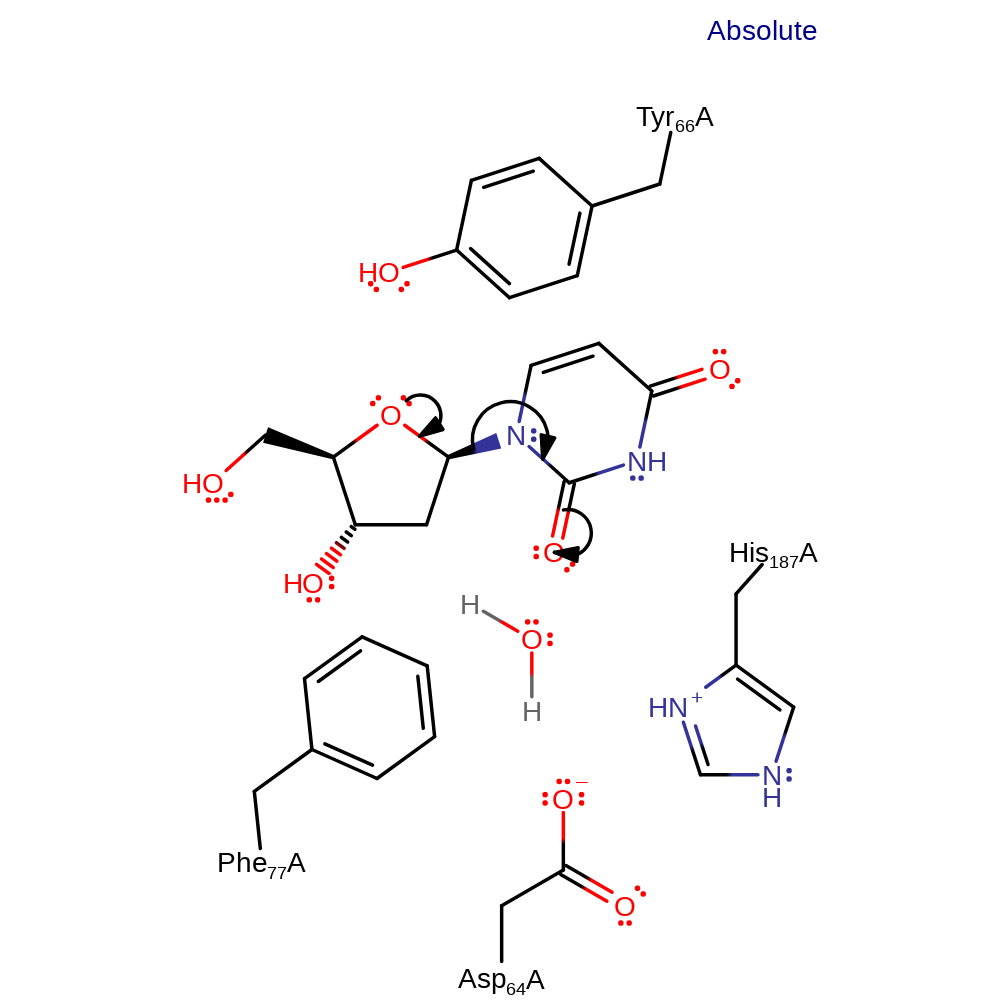
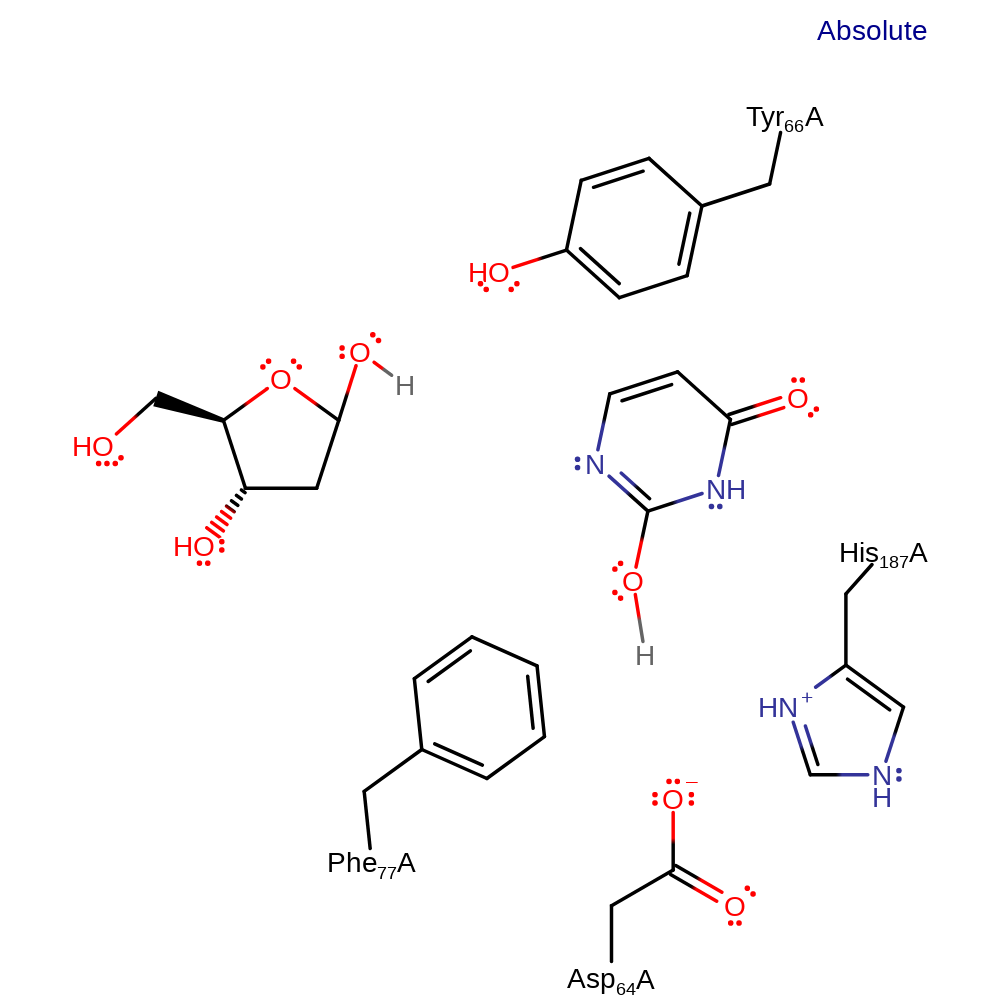
Step 1. The structural constraints imposed upon the uridine deoxyribose ring by Tyr 66 and Phe 77 induce unimolecular elimination to form a transient oxonium species and an anionic precursor of uracil
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr66A | steric role |
| His187A | hydrogen bond donor |
| Phe77A | steric role |
| Asp64A | electrostatic stabiliser |
Chemical Components
ingold: intramolecular elimination, intermediate formation, overall reactant used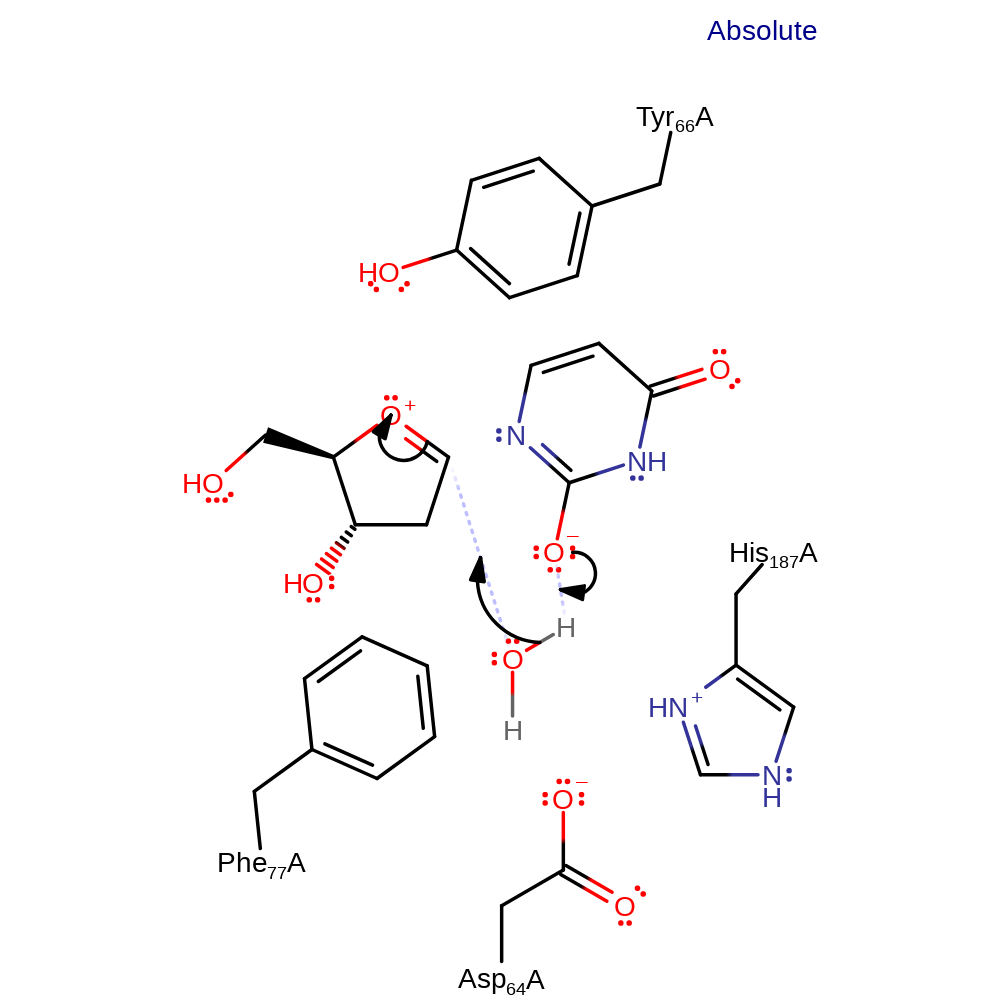
Step 2. The transient oxonium species is attacked by water, forming the 1-alpha hydroxy group of the D-ribose product. The anionic intermediate is stabilised though hydrogen bonding to His187, and is then protonated from the attacking water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr66A | activator, steric role |
| His187A | hydrogen bond donor, electrostatic stabiliser |
| Phe77A | activator, steric role |
| Asp64A | activator |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall product formed, native state of enzyme regenerated, intermediate terminated, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
tautomerisation (not keto-enol), reaction occurs outside the enzymeIntroduction
This proposal represents the classical acid/base proposal carried out by a cationic His-187 and an absolutely conserved Asp-64. Here the peptide carbonyl and side-chain carboxyl of Asp-64 activate a water molecule that attacks a weakened glycosylic bond. Destabilisation of the N1−C1‘ bond is brought about by distortion or protonation of the uracil O2 by His-187 Nε2.
Catalytic Residues Roles
| UniProt | PDB* (1eug) | ||
| Tyr66 | Tyr66A | The residue acts to distort the DNA substrate towards the transition state conformation through steric interactions with the pucker ring of the uridine deoxyribose. The residue is also implicated in preventing the binding of thymine within the active site. | steric role |
| Phe77 | Phe77A | The residue acts to distort the DNA substrate towards the transition state conformation by steric and pi stacking interactions with the pucker ring of the uridine deoxyribose. | steric role |
| His187 | His187A | The residue hydrogen bonds to the developing anion within the transition state, lowering its energy. | hydrogen bond donor, electrostatic stabiliser |
| Asp64 | Asp64A | Acts as a general acid/base. | proton acceptor, electrostatic stabiliser, proton donor |
Chemical Components
intermediate formation, overall reactant used, bimolecular nucleophilic substitution, overall product formed, proton transfer, assisted tautomerisation (not keto-enol), native state of enzyme regeneratedReferences
- Shroyer MJ et al. (1999), Biochemistry, 38, 4834-4845. Mutation of an active site residue in Escherichia coli uracil-DNA glycosylase: effect on DNA binding, uracil inhibition and catalysis. DOI:10.1021/bi982986j. PMID:10200172.

Step 1. Asp64 abstracts a proton from the catalytic water molecule. The activated hydroxide then attacks the anomeric carbon in a nucleophilic substitution reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp64A | electrostatic stabiliser |
| Tyr66A | steric role |
| His187A | hydrogen bond donor |
| Phe77A | steric role |
| Asp64A | proton acceptor |
Chemical Components
intermediate formation, overall reactant used, ingold: bimolecular nucleophilic substitution, overall product formed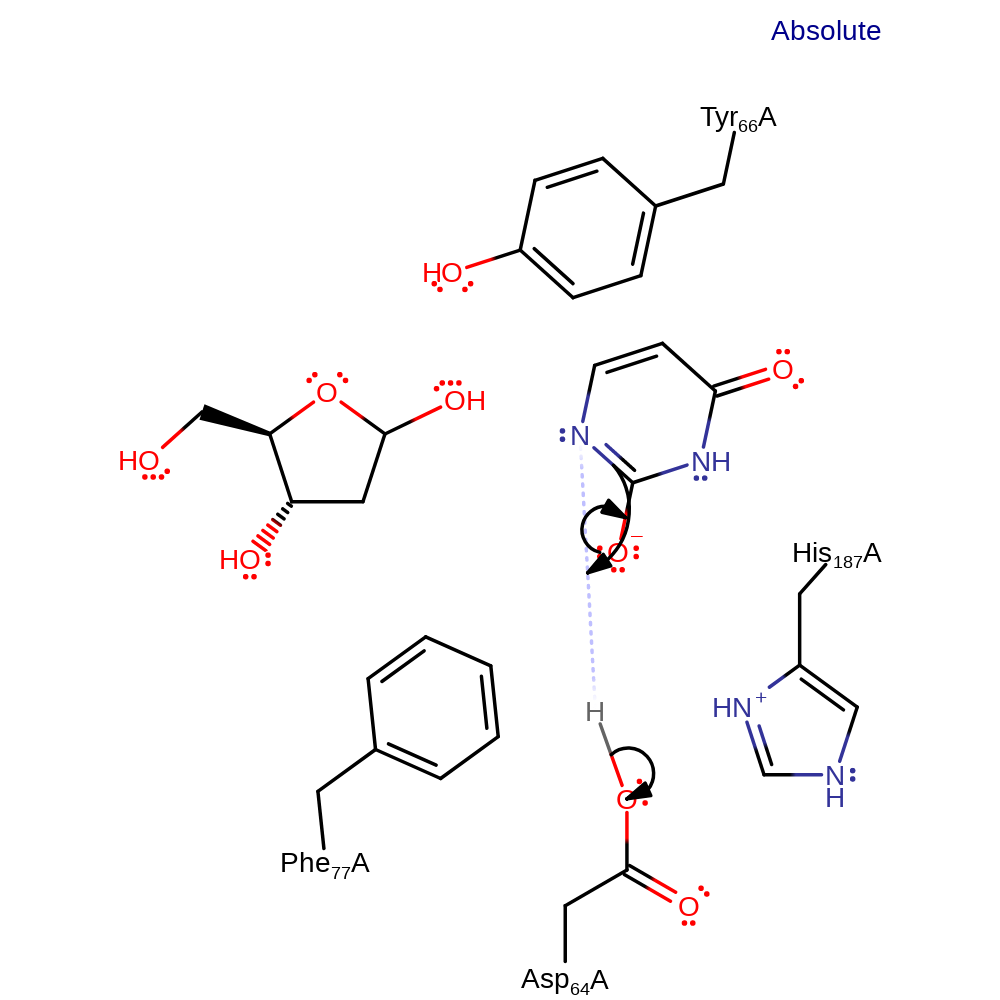
Step 2. The intermediate abstracts a proton from Asp64 to regenerate the active site and form the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr66A | steric role |
| Phe77A | steric role |
| His187A | electrostatic stabiliser |
| Asp64A | proton donor |
Chemical Components
proton transfer, overall product formed, assisted tautomerisation (not keto-enol), native state of enzyme regeneratedIntroduction
The imidazole group of His-187 catalyses a direct nucleophilic attack on the N1−C1‘ glycosylic bond of uracil and a second nucleophilic attack of a water molecule provides H- and OH-group addition to the N1 and C1‘ atoms, respectively.
Catalytic Residues Roles
| UniProt | PDB* (1eug) | ||
| Tyr66 | Tyr66A | The residue acts to distort the DNA substrate towards the transition state conformation through steric interactions with the pucker ring of the uridine deoxyribose. The residue is also implicated in preventing the binding of thymine within the active site | steric role |
| Phe77 | Phe77A | The residue acts to distort the DNA substrate towards the transition state conformation by steric and pi stacking interactions with the pucker ring of the uridine deoxyribose. | steric role |
| His187 | His187A | Acts as a nucleophile and forms a covalent intermediate with the sugar ring of DNA. | covalently attached, nucleofuge, nucleophile |
| Asp64 | Asp64A | Stabilising and activating the catalytic histidine and water molecule. | activator, electrostatic stabiliser, increase acidity |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, intermediate formation, enzyme-substrate complex formation, proton transfer, overall product formed, native state of enzyme regenerated, enzyme-substrate complex cleavageReferences
- Mol CD et al. (1995), Cell, 80, 869-878. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. PMID:7697717.
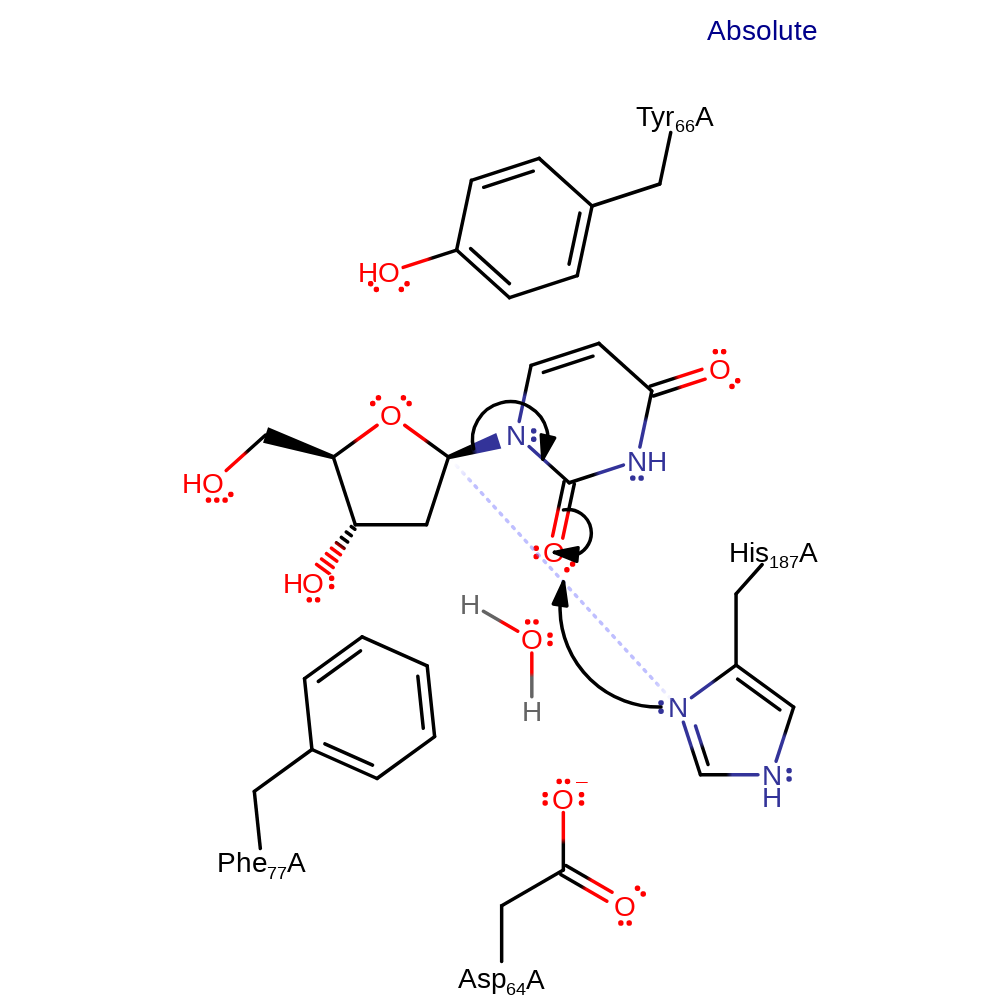
Step 1. His187 initiates a nucleophilic attack on the anomeric carbon of the DNA base.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe77A | steric role |
| Tyr66A | steric role |
| Asp64A | electrostatic stabiliser |
| His187A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, intermediate formation, enzyme-substrate complex formation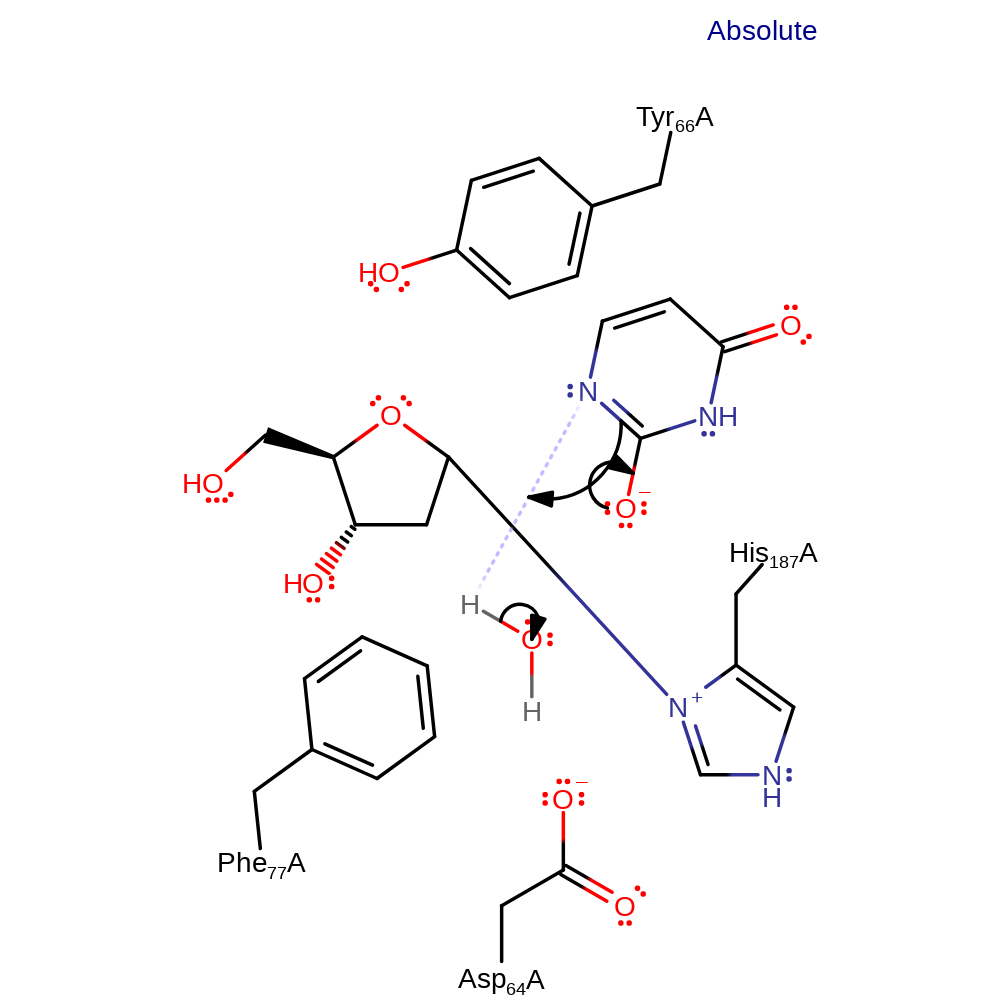
Step 2. The uracil undergoes tautomerisation and abstracts a proton from the catalytic water molecule, forming one of the final products and activating the water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp64A | increase acidity |
| Tyr66A | steric role |
| Phe77A | steric role |
| His187A | covalently attached |
Chemical Components
proton transfer, overall reactant used, overall product formed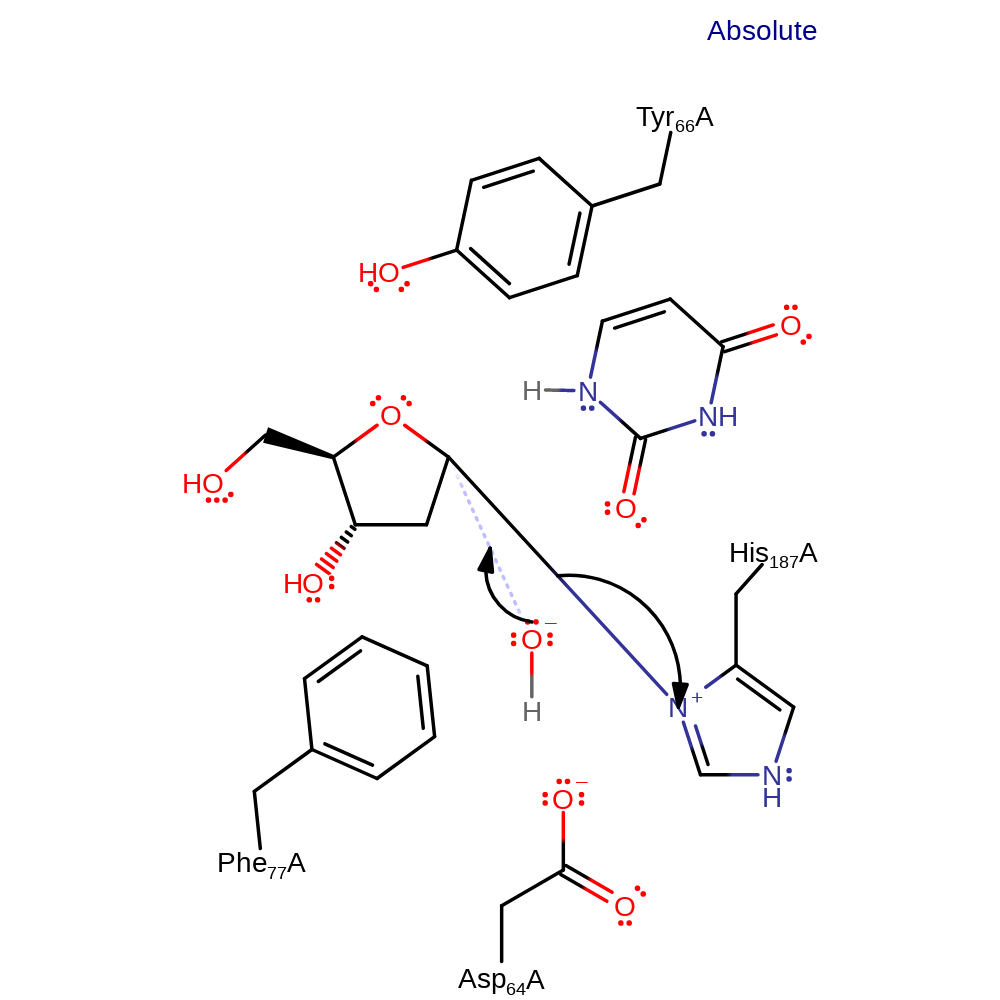
Step 3. The activated water then attacks the anomeric carbon of the enzyme bound sugar. This forms the final product and regenerates the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp64A | activator |
| Tyr66A | steric role |
| Phe77A | steric role |
| His187A | nucleofuge |
Chemical Components
ingold: bimolecular nucleophilic substitution, native state of enzyme regenerated, overall product formed, enzyme-substrate complex cleavageIntroduction
This proposal represents the second acid/base proposal carried out by a neutral His-187 and an absolutely conserved Asp-64. Here His-187 activates a water molecule that attacks a weakened glycosylic bond.
Catalytic Residues Roles
| UniProt | PDB* (1eug) | ||
| Tyr66 | Tyr66A | The residue acts to distort the DNA substrate towards the transition state conformation through steric interactions with the pucker ring of the uridine deoxyribose. The residue is also implicated in preventing the binding of thymine within the active site. | steric role |
| Phe77 | Phe77A | The residue acts to distort the DNA substrate towards the transition state conformation by steric and pi stacking interactions with the pucker ring of the uridine deoxyribose. | steric role |
| His187 | His187A | Acts as a general acid/base. | proton acceptor, proton donor |
| Asp64 | Asp64A | Activates the catalytic water molecule. | electrostatic stabiliser, increase acidity |
Chemical Components
overall product formed, bimolecular nucleophilic substitution, overall reactant used, intermediate formation, native state of enzyme regenerated, assisted tautomerisation (not keto-enol), proton transferReferences
- Drohat AC et al. (2014), Org Biomol Chem, 12, 8367-8378. Mechanisms for enzymatic cleavage of the N-glycosidic bond in DNA. DOI:10.1039/c4ob01063a. PMID:25181003.
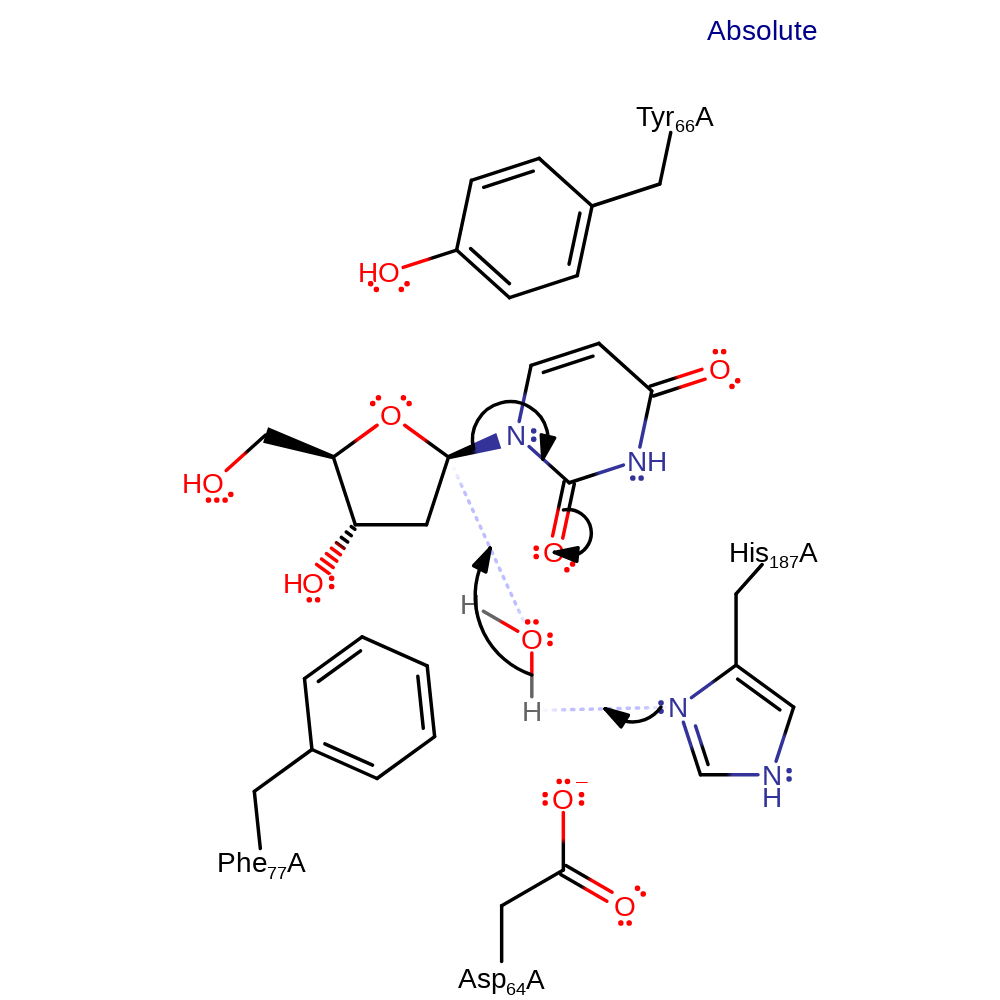
Step 1. His-187 abstracts a proton from the catalytic water molecule. The activated hydroxide then attacks the anomeric carbon in a nucleophilic substitution reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe77A | steric role |
| Tyr66A | steric role |
| Asp64A | electrostatic stabiliser |
| Asp64A | increase acidity |
| His187A | proton acceptor |
Chemical Components
overall product formed, ingold: bimolecular nucleophilic substitution, overall reactant used, intermediate formation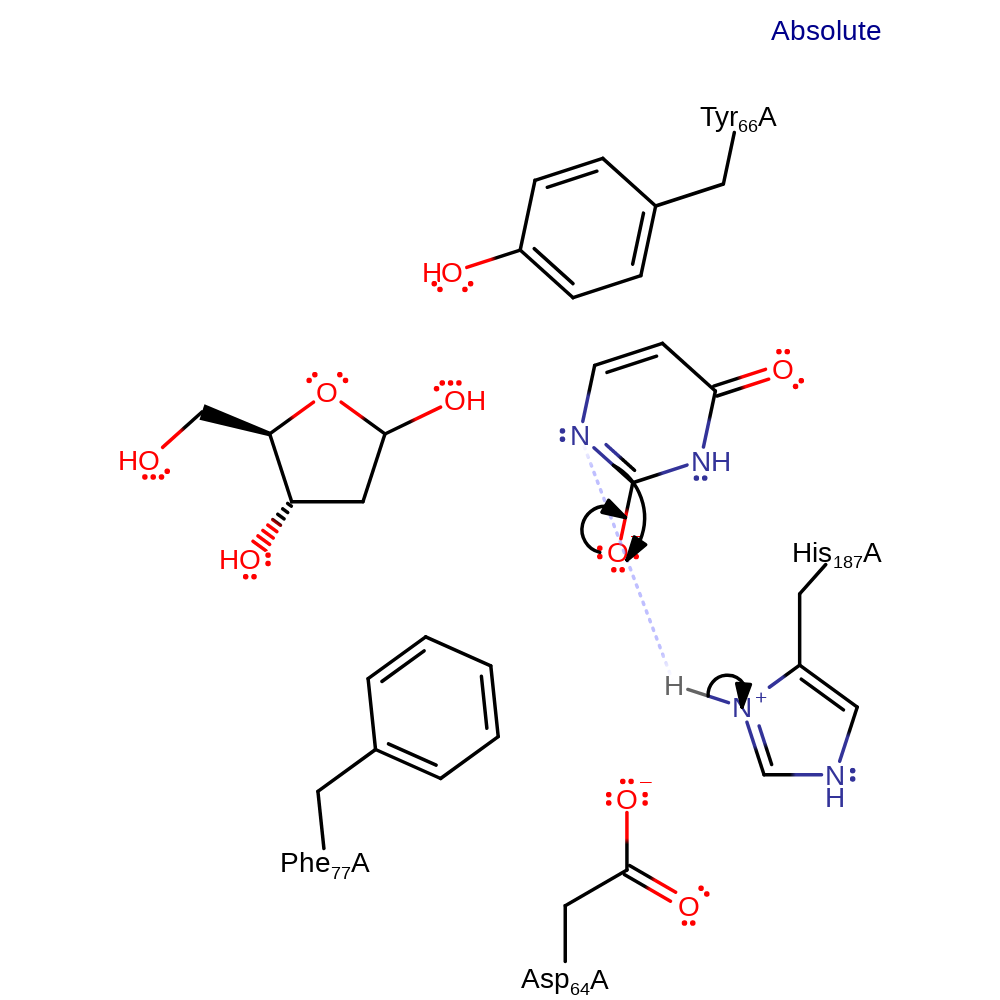
Step 2. The intermediate abstracts a proton from His-187 to regenerate the active site and form the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe77A | steric role |
| Tyr66A | steric role |
| His187A | proton donor |

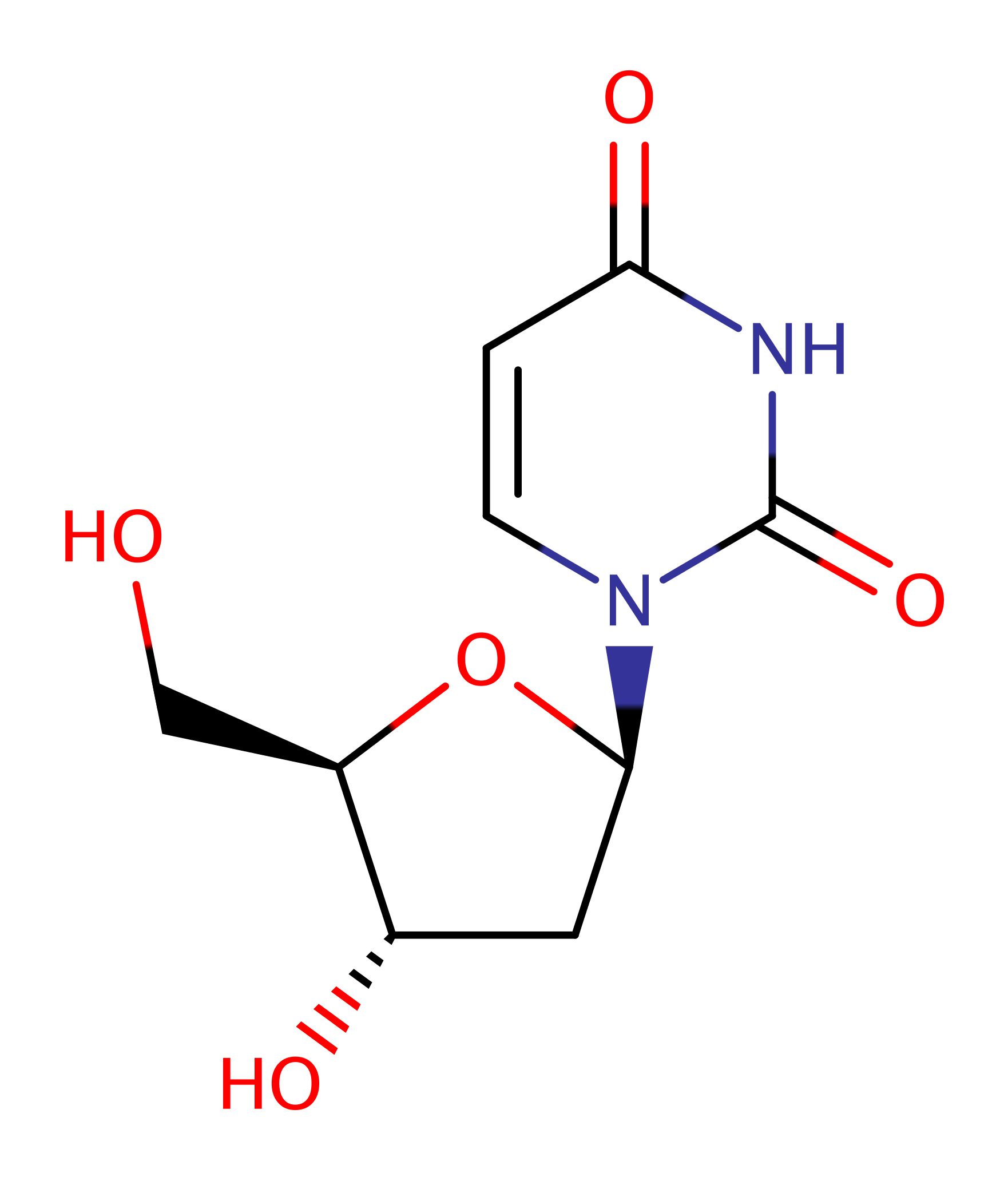
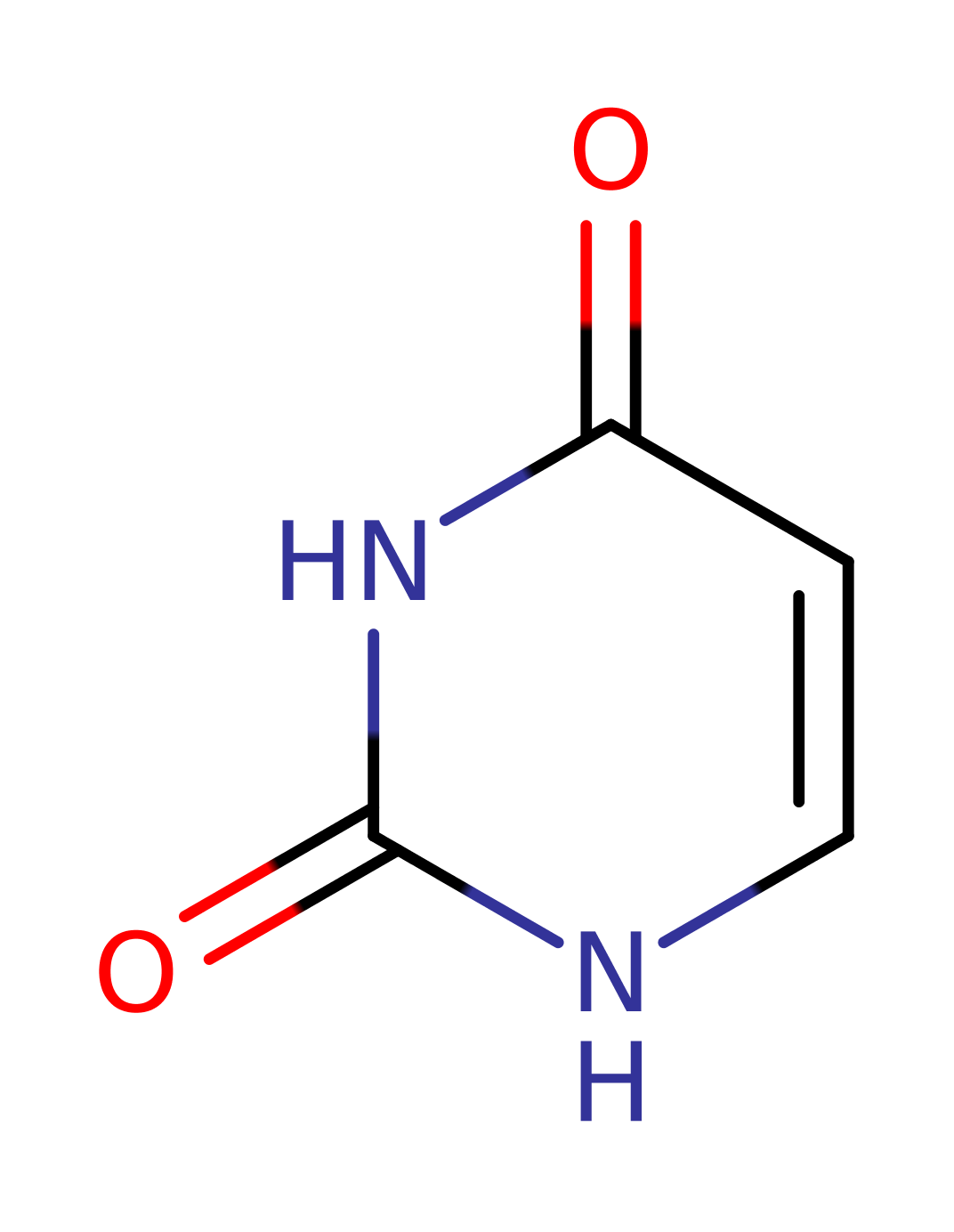
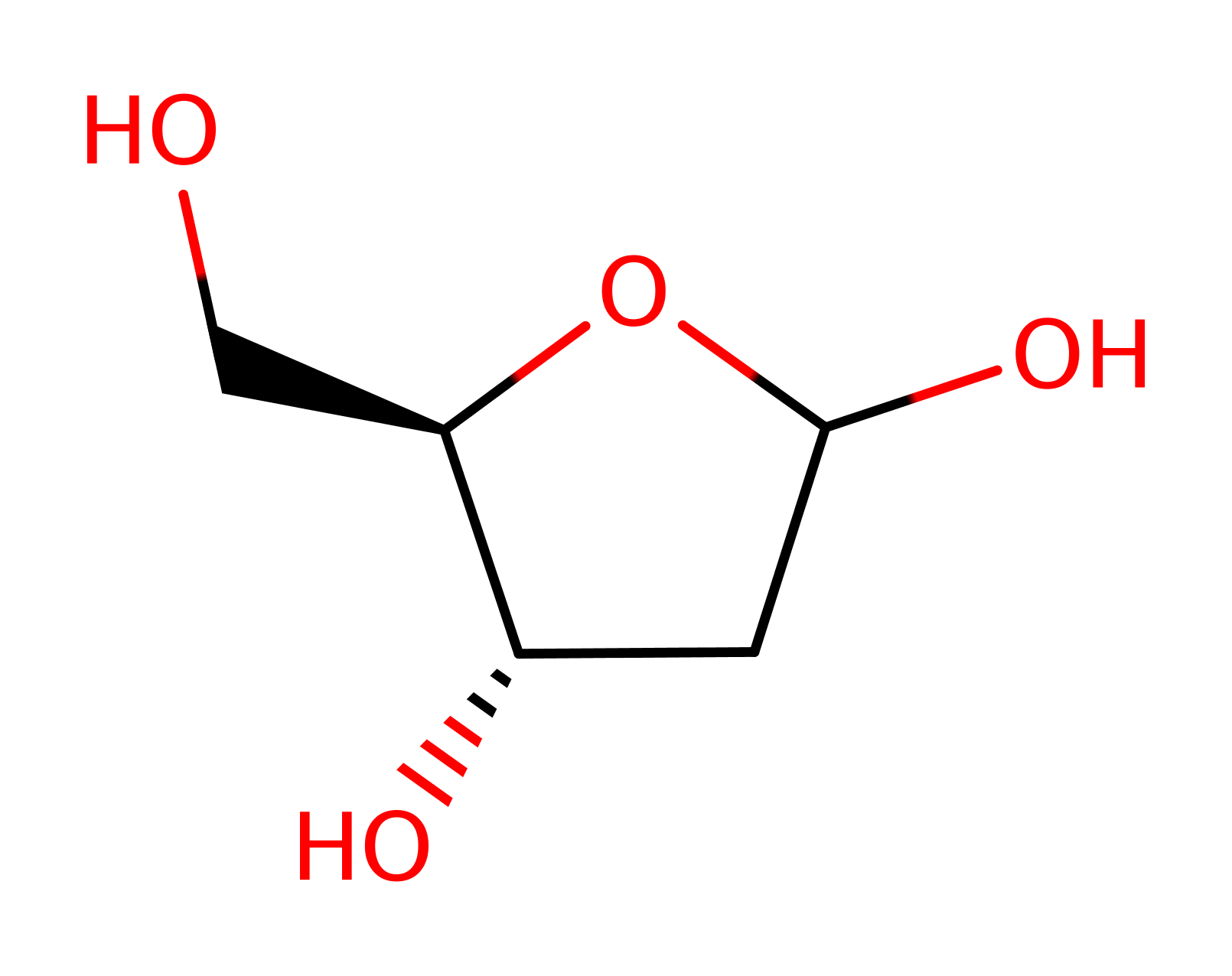
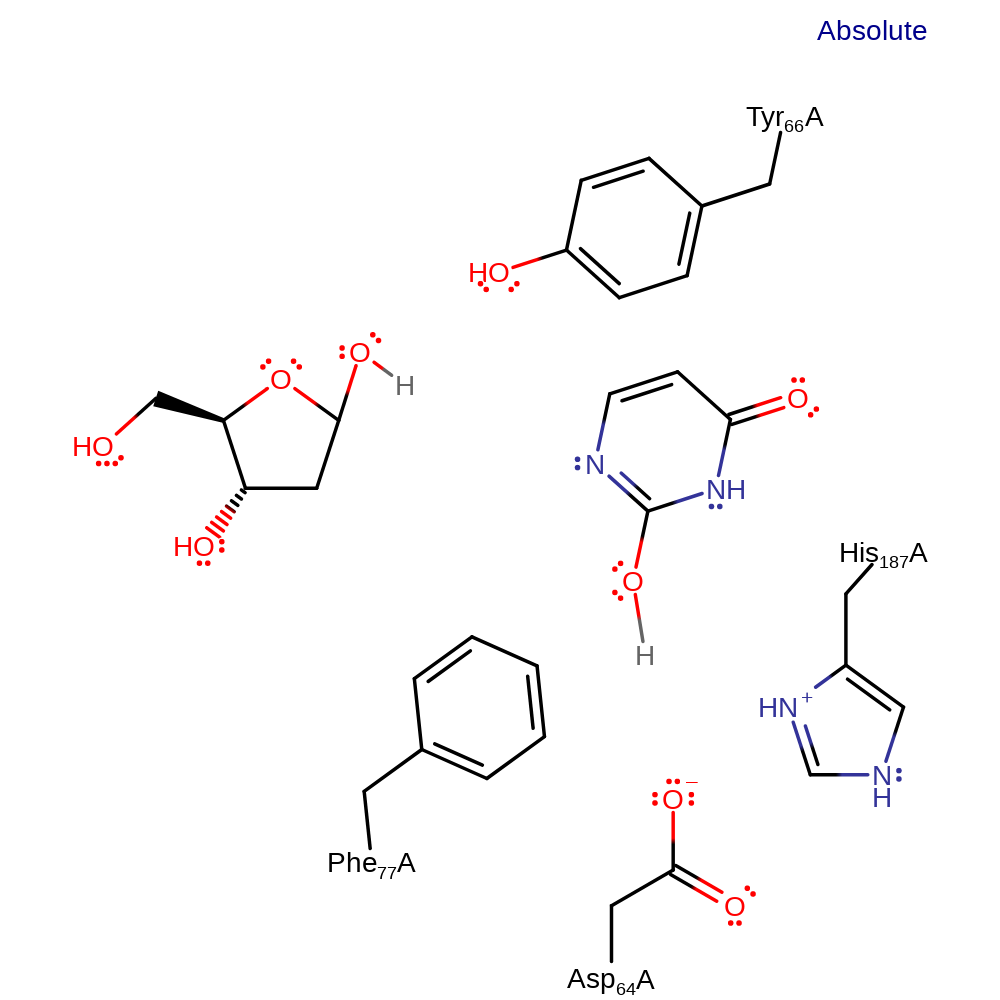
 Download:
Download:  Download:
Download: 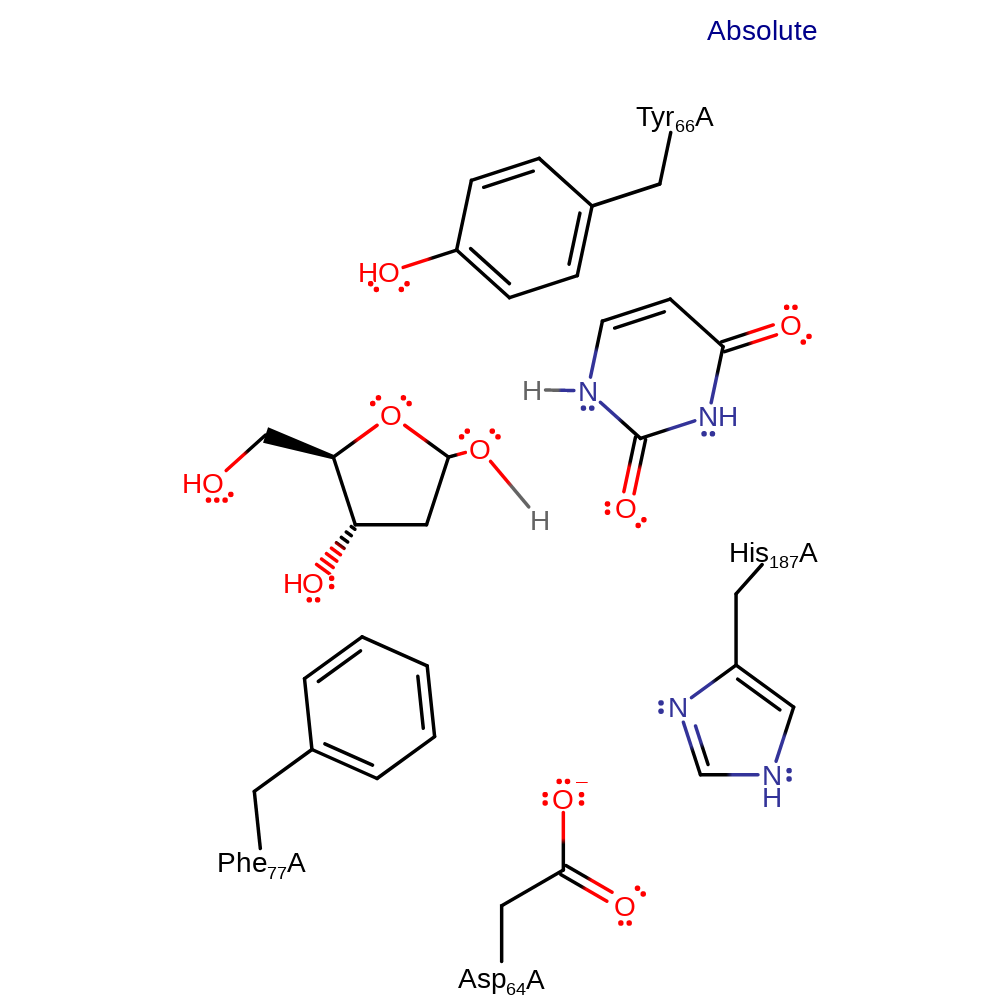 Download:
Download: 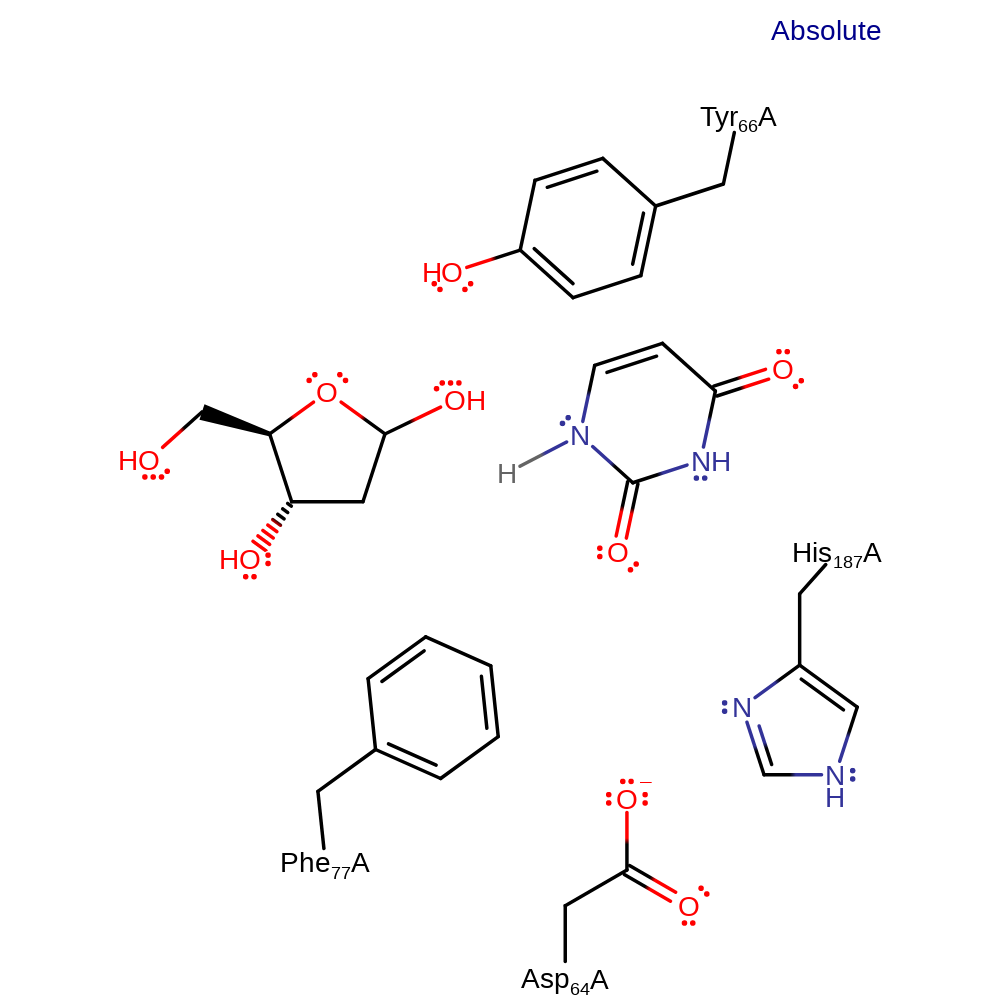 Download:
Download: