3-dehydroquinate dehydratase (type I)
3-dehydroquinate dehydratase catalyses the third step in the biosynthesis of chorismate within the Shikimate pathway which synthesises aromatic compounds as well as in the degradative quinate pathway. It is a type I dehydroquinase which catalyses a cis-dehydration of the hexane ring of 3-dehydroquinate via a covalent imine intermediate (unlike the type II dehydroquinase which catalyses a trans-dehydration via an enolate intermediate). Type I dehydroquinases use a Schiff base mechanism. The pathway is essential in microorganisms and plants for the biosynthesis of compounds such as folate, ubiquinone and aromatic amino acids. The absence of this pathway in animals makes it an attractive target for antimicrobial agents.
Reference Protein and Structure
- Sequence
-
P24670
 (4.2.1.10)
(4.2.1.10)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Salmonella enterica subsp. enterica serovar Typhi (Bacteria)

- PDB
-
1qfe
- THE STRUCTURE OF TYPE I 3-DEHYDROQUINATE DEHYDRATASE FROM SALMONELLA TYPHI
(2.1 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 1qfe)
(see all for 1qfe)
Enzyme Reaction (EC:4.2.1.10)
Enzyme Mechanism
Introduction
His143 is thought to play a part as a general acid in the formation of a Schiff base: a covalent adduct between the substrate and Lys170 of the enzyme. The role of the Schiff base is to act as an electron sink . It may also play a role in distorting the carbocyclic ring of dehydroquinate to render it more reactive. His143 is then thought to play a role in proton abstraction. Glu86 is positioned to interact with His143 and orientate it in a manner reminiscent of the serine proteases to allow it to act as a general base and abstract the C2 proton. However it is worth noting that recent research has put some doubt on this role. Any attack on the substrate from below is prevented by a beta-hairpin so only cis-elimination is possible.
Catalytic Residues Roles
| UniProt | PDB* (1qfe) | ||
| His143 | His143A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Lys170 | Lys170A | Acts as the catalytic nucleophile. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleophile, proton acceptor, proton donor, nucleofuge, electron pair acceptor, electron pair donor |
| Glu86 | Glu86A | Acts as a general acid/base | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
Chemical Components
proton transfer, proton relay, bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, inferred reaction step, unimolecular elimination by the conjugate base, dehydration, enzyme-substrate complex cleavage, intermediate collapse, schiff base formed, assisted tautomerisation (not keto-enol), overall product formed, intramolecular elimination, intermediate terminated, native state of enzyme regeneratedReferences
- Leech AP et al. (1998), J Biol Chem, 273, 9602-9607. Re-evaluating the Role of His-143 in the Mechanism of Type I Dehydroquinase from Escherichia coli Using Two-dimensional 1H,13C NMR. DOI:10.1074/jbc.273.16.9602. PMID:9545291.
- Tizón L et al. (2015), Org Biomol Chem, 13, 706-716. Irreversible covalent modification of type I dehydroquinase with a stable Schiff base. DOI:10.1039/c4ob01782j. PMID:25370445.
- Light SH et al. (2014), Biochemistry, 53, 872-880. Crystal Structures of Type I Dehydroquinate Dehydratase in Complex with Quinate and Shikimate Suggest a Novel Mechanism of Schiff Base Formation. DOI:10.1021/bi4015506. PMID:24437575.
- Maneiro M et al. (2014), Biochem J, 462, 415-424. Insights into substrate binding and catalysis in bacterial type I dehydroquinase. DOI:10.1042/bj20140614. PMID:24957267.
- Light SH et al. (2013), Protein Sci, 22, 418-424. Reassessing the type I dehydroquinate dehydratase catalytic triad: Kinetic and structural studies of Glu86 mutants. DOI:10.1002/pro.2218. PMID:23341204.
- Lee WH et al. (2002), Acta Crystallogr D Biol Crystallogr, 58, 798-804. Comparison of different crystal forms of 3-dehydroquinase from Salmonella typhi and its implication for the enzyme activity. PMID:11976491.
- Gourley DG et al. (1999), Nat Struct Biol, 6, 521-525. The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction. DOI:10.1038/9287. PMID:10360352.
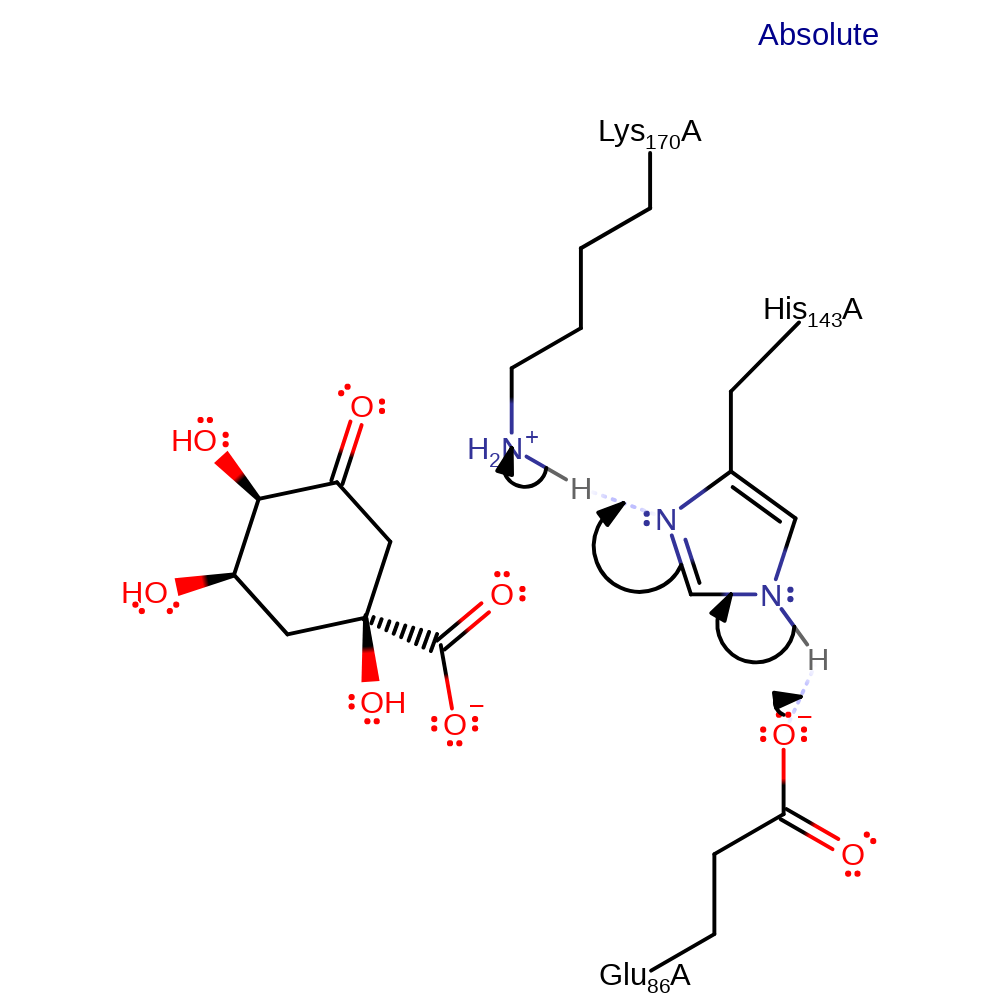
Step 1. Glu86 deprotonates His143, which deprotonates Lys170, activating it.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu86A | hydrogen bond acceptor |
| His143A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Lys170A | hydrogen bond donor, proton donor |
| Glu86A | proton acceptor |
| His143A | proton donor, proton acceptor |
Chemical Components
proton transfer, proton relay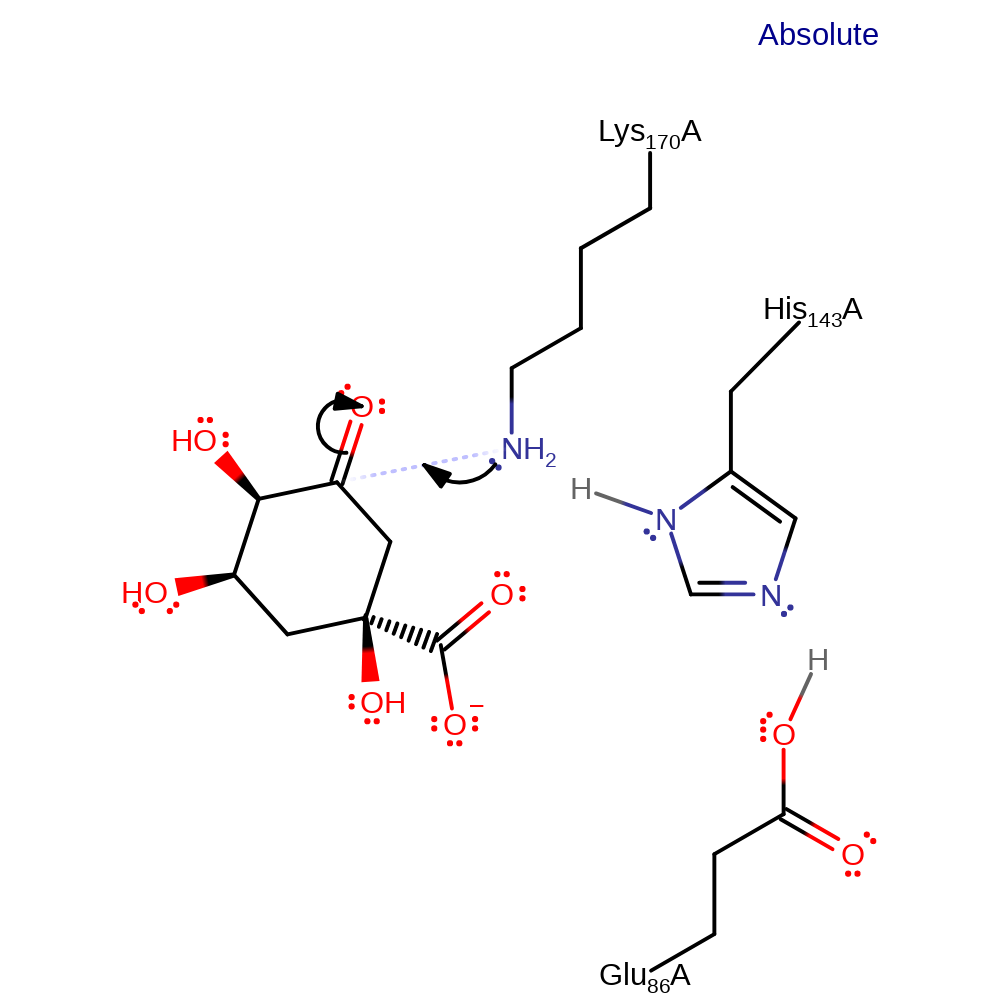
Step 2. Lys170 attacks the carbonyl carbon of the substrate in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu86A | hydrogen bond donor |
| His143A | hydrogen bond acceptor |
| Lys170A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation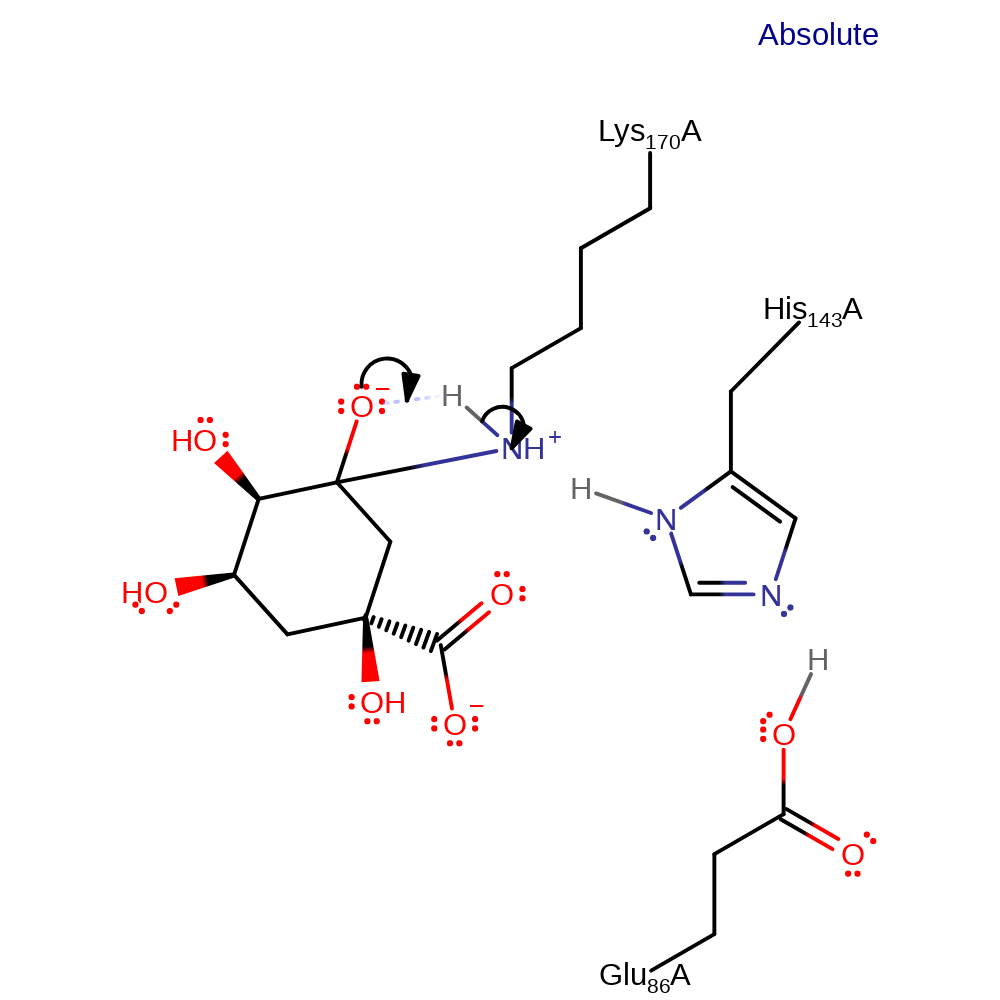
Step 3. A proton is transferred from the covalently attached lysine to the newly formed hydroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His143A | hydrogen bond acceptor |
| Glu86A | hydrogen bond donor |
| Lys170A | covalently attached, hydrogen bond donor |
| Lys170A | proton donor |
Chemical Components
proton transfer, intermediate formation, inferred reaction step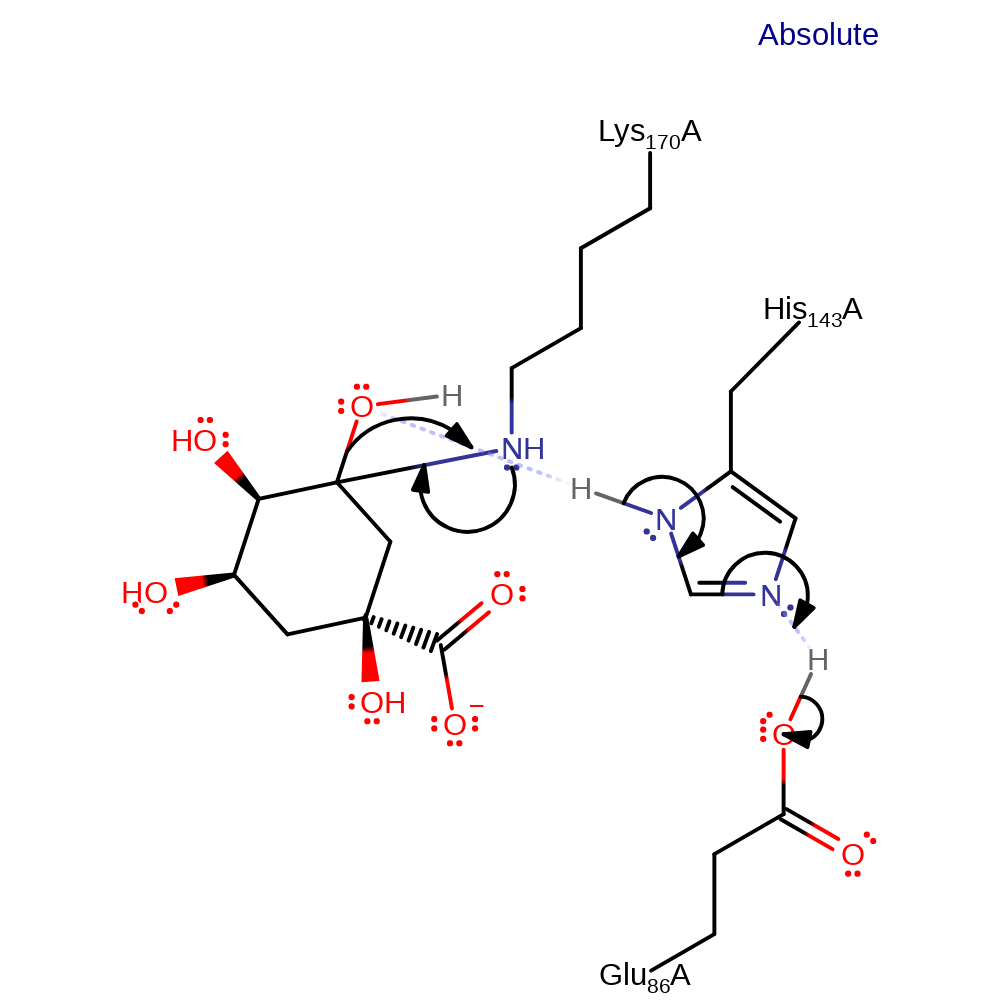
Step 4. Lys170 initiates an elimination of water (which obtains its proton from His143, which deprotonates Glu86) forming the Schiff base intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His143A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Lys170A | covalently attached |
| Glu86A | hydrogen bond donor, proton donor |
| His143A | proton acceptor, proton donor |
| Lys170A | electron pair donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, dehydration, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation, proton relay, schiff base formed, inferred reaction step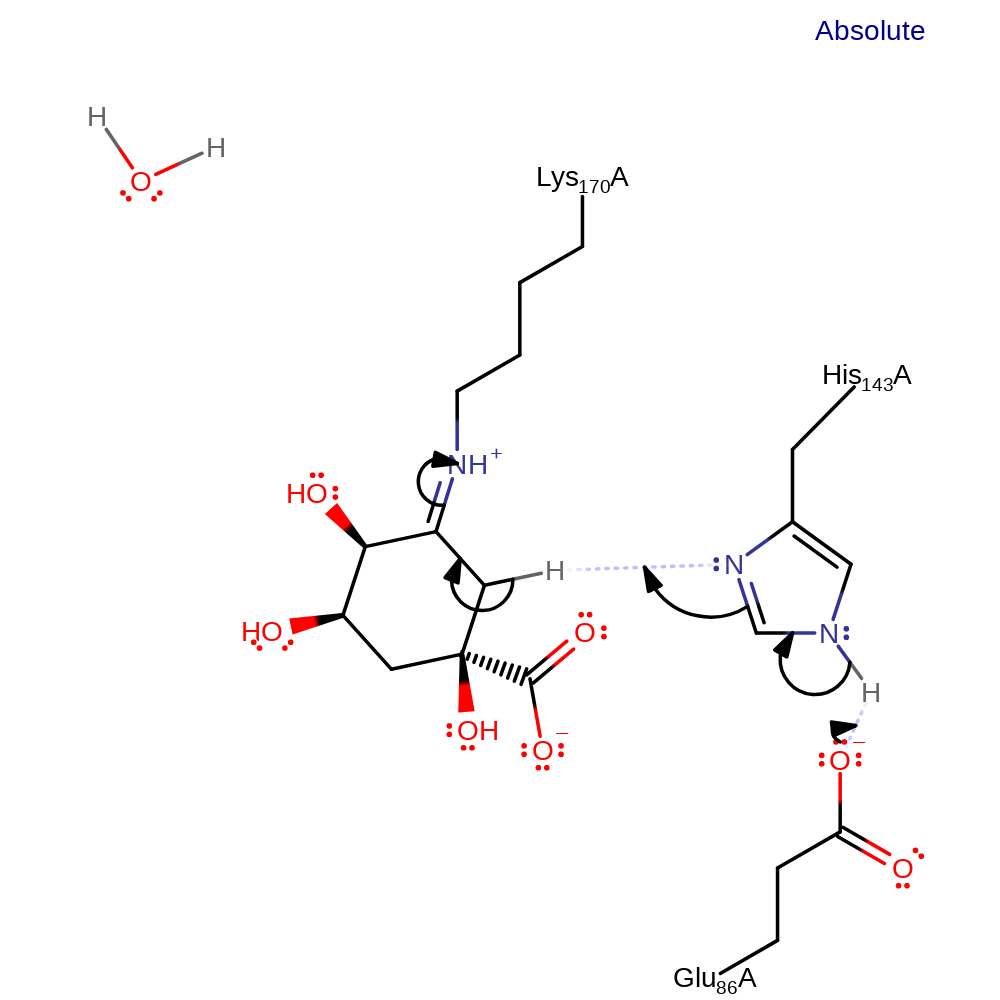
Step 5. Glu86 deprotonates His143, which deprotonates the intermediate at the carbon adjacent to the covalently bound lysine which acts as an electron sink.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu86A | hydrogen bond acceptor |
| His143A | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Lys170A | covalently attached |
| His143A | proton donor, proton acceptor |
| Glu86A | proton acceptor |
| Lys170A | electron pair acceptor |
Chemical Components
assisted tautomerisation (not keto-enol), proton transfer, intermediate formation, proton relay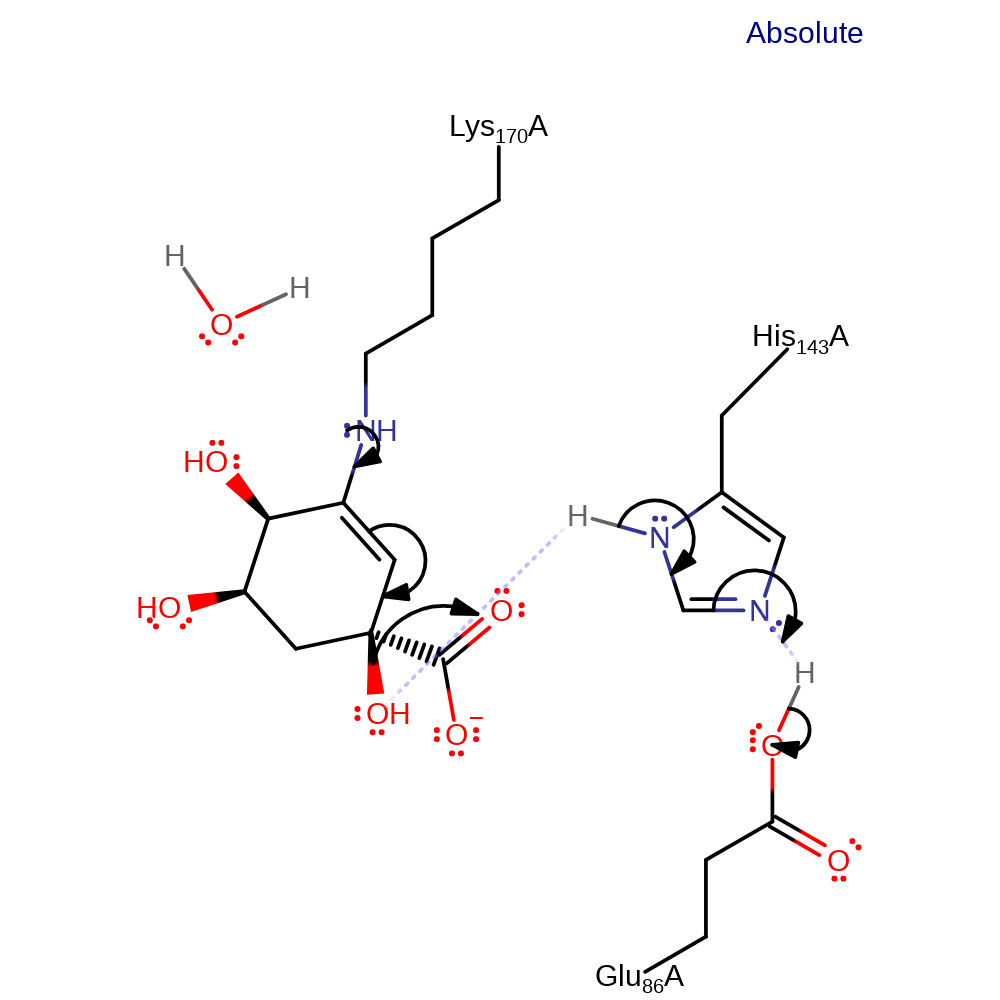
Step 6. Lys170 donates its lone pair of electrons back into the ring, initiating a double bond rearrangement and elimination of water, which obtains its proton from His143, which deprotonates Glu86.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu86A | hydrogen bond donor |
| His143A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Lys170A | covalently attached |
| Glu86A | proton donor |
| His143A | proton acceptor, proton donor |
| Lys170A | electron pair donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, overall product formed, dehydration, enzyme-substrate complex cleavage, intermediate formation, intermediate collapse, proton relay
Step 7. Glu86 deprotonates His143, which deprotonates water, which then attacks the carbon to which Lys170 is covalently attached.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His143A | proton relay, hydrogen bond acceptor, hydrogen bond donor |
| Glu86A | hydrogen bond acceptor |
| Lys170A | covalently attached |
| His143A | proton acceptor, proton donor |
| Glu86A | proton acceptor |
| Lys170A | electron pair acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, enzyme-substrate complex formation, intermediate formation, proton relay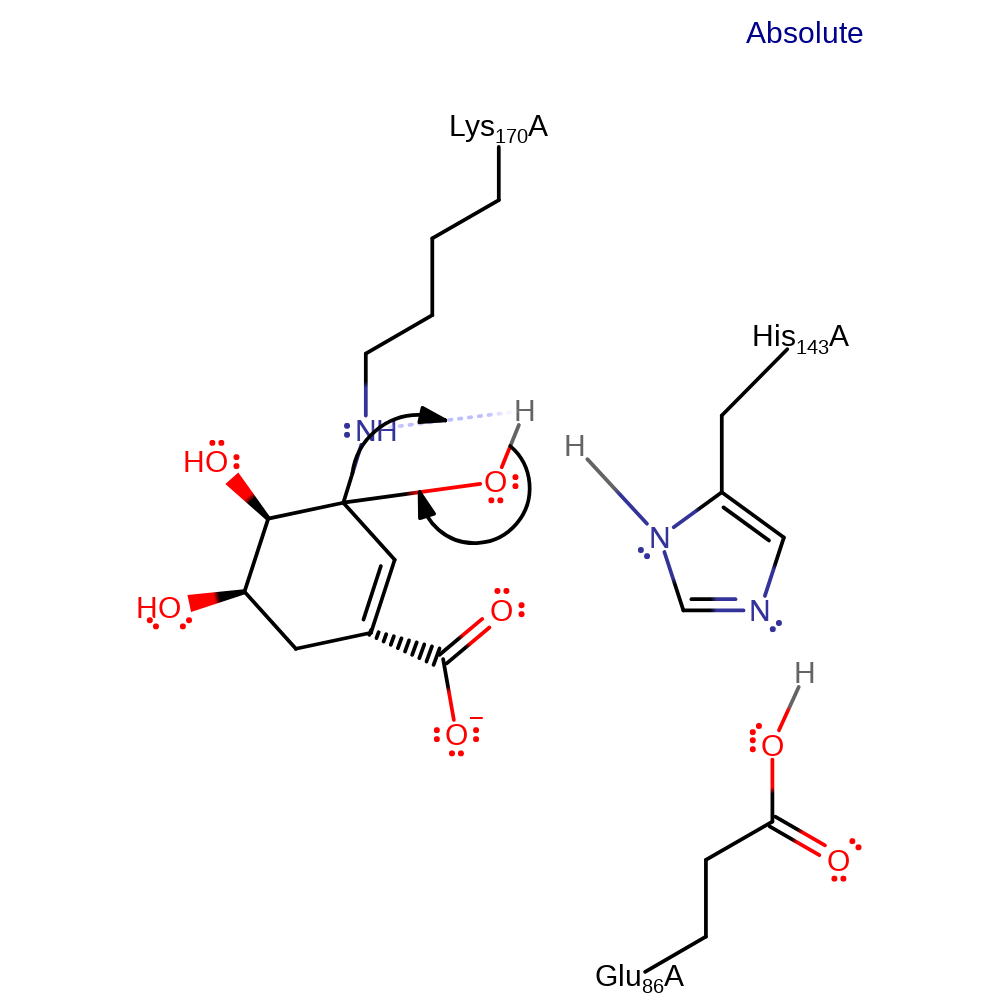
Step 8. Lys170 deprotonates the hydroxide, which causes Lys170 to be eliminated and the product to be formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu86A | hydrogen bond donor |
| His143A | hydrogen bond acceptor, hydrogen bond donor |
| Lys170A | hydrogen bond acceptor, proton acceptor, nucleofuge |
Chemical Components
ingold: intramolecular elimination, proton transfer, enzyme-substrate complex cleavage, overall product formed, intermediate collapse, intermediate terminated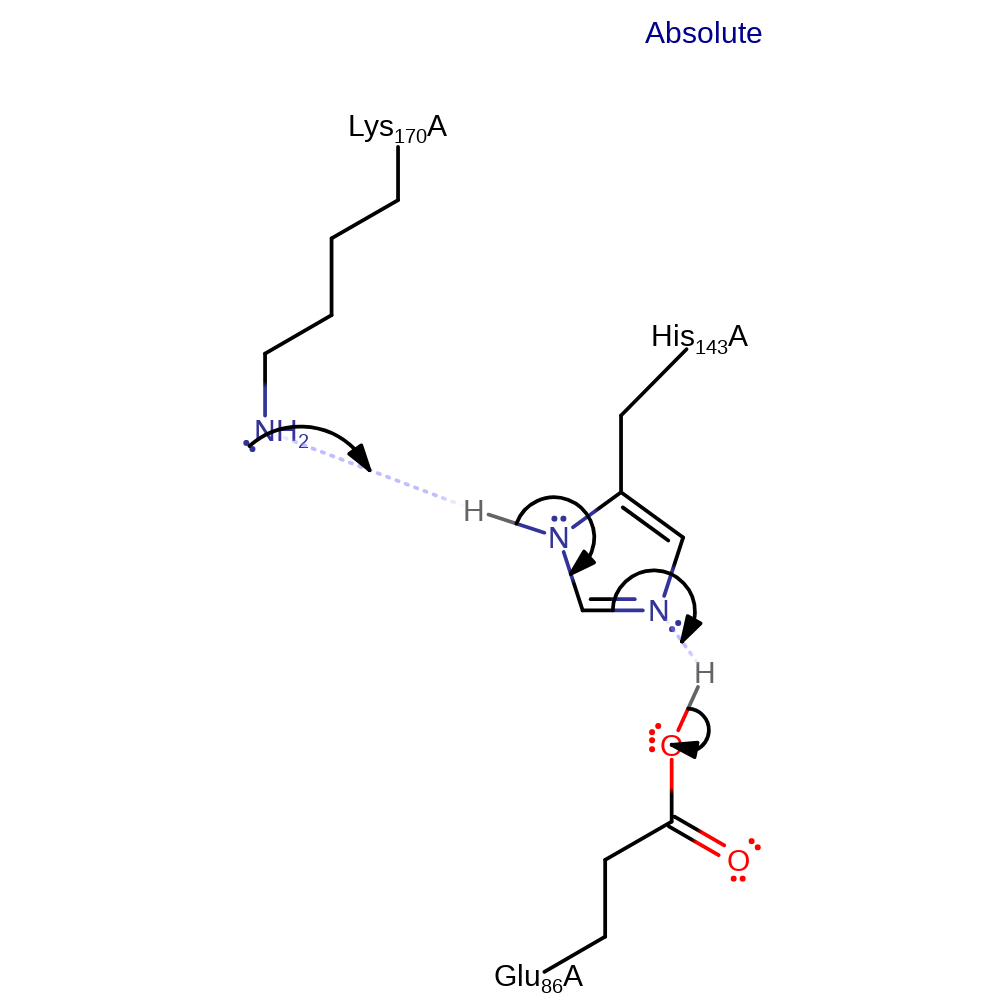
Step 9. Lys170 deprotonates His143, which deprotonates Glu86 in an inferred step that returns the enzyme to its starting state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu86A | hydrogen bond donor |
| His143A | hydrogen bond donor, hydrogen bond acceptor, proton relay |
| Lys170A | hydrogen bond acceptor |
| His143A | proton acceptor |
| Glu86A | proton donor |
| Lys170A | proton acceptor |
| His143A | proton donor |



 Download:
Download: