Malate synthase
The discovery of malate synthase proved to be the missing link in closing the tricarboxylic acid cycle, also known as the glyoxylate cycle. Together with isocitrate lyase, malate synthase allows the utilisation of two carbon compounds that would otherwise be wasted. Firstly isocitrate lyase catalyses the cleavage of isocitrate to succinate and glyoxylate (the citric acid cycle would otherwise convert isocitrate to succinate and two molecules of carbon dioxide). Malate synthase then catalyses the Claisen condensation of glyoxylate with an acetyl group from acetyl-CoA to form a malyl-CoA intermediate. This is subsequently hydrolysed, producing malate to replenish the pool of citric-acid-cycle intermediates.
Reference Protein and Structure
- Sequence
-
P37330
 (2.3.3.9)
(2.3.3.9)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1d8c
- MALATE SYNTHASE G COMPLEXED WITH MAGNESIUM AND GLYOXYLATE
(2.0 Å)



- Catalytic CATH Domains
-
3.20.20.360
 1.20.1220.12
1.20.1220.12  (see all for 1d8c)
(see all for 1d8c)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:2.3.3.9)
Enzyme Mechanism
Introduction
The electrophilic substrate is polarised for nucleophilic attack by hydrogen bonds and nearby positive charges, for example the Mg(II) ion coordinated by Glu427 and Asp455. Asp631 and Arg338 carry out the actual catalytic steps: Firstly Asp631 acts as a catalytic base to deprotonate the terminal acetyl group of acetyl CoA forming the enolate intermediate. The negative charge formed on the enolate oxygen is stabilised by the adjacent positive Arg338. The positively charged side chain is itself stabilised by interactions with the nearby residues Asp270 and Glu272. This high energy intermediate then mediates a nucleophilic attack on the 2-carbon of glyoxylate. The resulting oxyanion is stabilised by the positively charged Mg(II) and Arg338 until product release when protons would become available from the solution or presumably activated water molecule which hydrolyses the thioester.
Catalytic Residues Roles
| UniProt | PDB* (1d8c) | ||
| Asp455, Glu427 | Asp455A, Glu427A | Bind Mg(II) ion. | metal ligand |
| Asp270, Glu272 | Asp270A, Glu272A | The residue is important in stabilising the positive charge of Arg338, which in turn acts to stabilise the enolate intermediate. | hydrogen bond acceptor, electrostatic stabiliser |
| Arg338 | Arg338A | The positively charged side chain stabilises the enolate intermediate. | hydrogen bond donor, electrostatic stabiliser |
| Asp631 | Asp631A | The residue acts as a general base towards acetyl-CoA, deprotonating the terminal acetyl group to form an enolate intermediate. At the end of the reaction, the initial carboxylate group of Asp631 is regenerated by removal of the residue's acquired proton by Arg338. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, overall reactant used, intermediate formation, aldol addition, bimolecular nucleophilic addition, bimolecular nucleophilic substitution, overall product formed, native state of enzyme regenerated, intermediate collapse, intermediate terminated, hydrolysisReferences
- Bracken CD et al. (2011), BMC Struct Biol, 11, 23-. Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G. DOI:10.1186/1472-6807-11-23. PMID:21569248.
- Huang HL et al. (2016), J Biol Chem, 291, 27421-27432. Mycobacterium tuberculosisMalate Synthase Structures with Fragments Reveal a Portal for Substrate/Product Exchange. DOI:10.1074/jbc.m116.750877. PMID:27738104.
- Lohman JR et al. (2008), Protein Sci, 17, 1935-1945. Atomic resolution structures ofEscherichia coliandBacillus anthracismalate synthase A: Comparison with isoform G and implications for structure-based drug discovery. DOI:10.1110/ps.036269.108. PMID:18714089.
- Anstrom DM et al. (2006), Protein Sci, 15, 2002-2007. The product complex ofM. tuberculosismalate synthase revisited. DOI:10.1110/ps.062300206. PMID:16877713.
- Smith CV et al. (2003), J Biol Chem, 278, 1735-1743. Biochemical and Structural Studies of Malate Synthase fromMycobacterium tuberculosis. DOI:10.1074/jbc.m209248200. PMID:12393860.
- Howard BR et al. (2000), Biochemistry, 39, 3156-3168. Crystal Structure ofEscherichia coliMalate Synthase G Complexed with Magnesium and Glyoxylate at 2.0 Å Resolution: Mechanistic Implications†,‡,§. DOI:10.1021/bi992519h. PMID:10715138.

Step 1. Asp631 deprotonates the methyl group of acetyl-CoA, which causes a rearrangement of the double bonds and the formation of an oxyanion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg338A | hydrogen bond donor, electrostatic stabiliser |
| Asp270A | electrostatic stabiliser, hydrogen bond acceptor |
| Asp631A | hydrogen bond acceptor |
| Glu272A | electrostatic stabiliser, hydrogen bond acceptor |
| Glu427A | metal ligand |
| Asp455A | metal ligand |
| Asp631A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, overall reactant used, intermediate formation
Step 2. The CoA intermediate rotates in the active site. The oxyanion collapses, initiating a nucleophilic attack on the carbonyl carbon of the oxo-acetic acid substrate in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg338A | electrostatic stabiliser, hydrogen bond donor |
| Asp270A | hydrogen bond acceptor, electrostatic stabiliser |
| Asp631A | electrostatic stabiliser, hydrogen bond donor |
| Glu272A | hydrogen bond acceptor, electrostatic stabiliser |
| Asp455A | metal ligand |
| Glu427A | metal ligand |
Chemical Components
aldol addition, ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation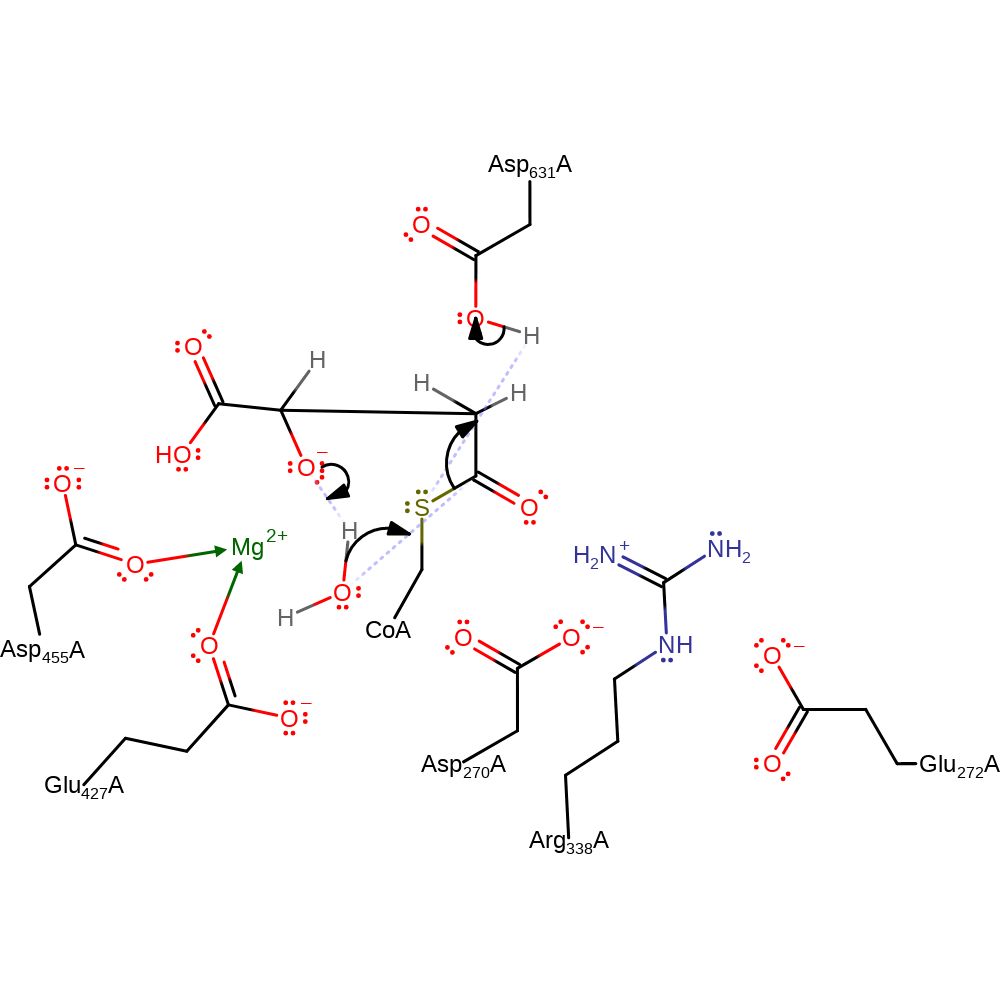
Step 3. The oxyanion deprotonates water, which initiates a nucleophilic attack on the carbonyl carbon adjacent to the sulfur of CoA in a substitution reaction, eliminating CoA with concomitant deprotonation of Asp631.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg338A | electrostatic stabiliser, hydrogen bond donor |
| Asp270A | hydrogen bond acceptor, electrostatic stabiliser |
| Asp631A | hydrogen bond donor |
| Glu272A | hydrogen bond acceptor, electrostatic stabiliser |
| Asp455A | metal ligand |
| Glu427A | metal ligand |
| Asp631A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, native state of enzyme regenerated, intermediate collapse, intermediate terminated, hydrolysisIntroduction
The mechanism starts with the Mg(II) ion octahedrally coordinated by the carboxylate side chains of conserved residues Glu434 and Asp462, and four water molecules as seen in the crystal structure 1N8I. Glyoxylate binds first displacing two of the metal-coordinated water molecules. After the binding of AcCoA, the conserved Asp633 is the catalytic base exhibiting the pKa value of 4.6 – 5.3, which abstracts a proton from AcCoA. This step is partially rate-limiting. In support of this, the analogous E. coli residue, Asp631, was mutated to Asn and exhibited no activity.
The resulting nucleophile attacks glyoxylate to form the malyl-CoA intermediate, which we draw as the alkoxide. The formation of the malyl-CoA intermediate is the first irreversible step.
The alkoxide serves to remove the proton from an adjacent metal-bound water, creating the hydroxide anion that attacks the carbonyl of the thioester intermediate.
The tetrahedral intermediate now decomposes with the generation of the two products. Here Arg339 acts as a catalytic acid to protonate the leaving group, the thiolate of CoA.
In the Mtb MS-CoA-malate structure, one of the ureido nitrogens is 3.6 Å from the thiol of CoASH. The analogous E. coli residue, Arg338 was mutated to Lys and exhibited only 6.6% of WT activity.
The ordered release of CoA followed by L-malate completes the catalytic cycle.
Catalytic Residues Roles
| UniProt | PDB* (1d8c) | ||
| Arg339, Asp633 | Arg339A, Asp633A | Acts as a general acid/base. | proton acceptor, electrostatic stabiliser, proton donor |
| Asp462, Glu434 | Asp462A, Glu434A | Forms part of the Mg(II) binding site. | metal ligand |
Chemical Components
proton transfer, bimolecular nucleophilic addition, bimolecular elimination, inferred reaction step, native state of enzyme regeneratedReferences
- Quartararo CE et al. (2011), Biochemistry, 50, 6879-6887. Kinetic and Chemical Mechanism of Malate Synthase fromMycobacterium tuberculosis. DOI:10.1021/bi2007299. PMID:21728344.

Step 1. Asp633 deprotonates the methyl group of CoA substrate, which initiates a nucleophilic attach on the pyruvate substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp462A | metal ligand |
| Glu434A | metal ligand |
| Arg339A | electrostatic stabiliser |
| Asp633A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition
Step 2. Mg(II) bound water is deprotonated by the oxyanion, initiating a nucleophilic attack on the carbonyl group of CoA.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu434A | metal ligand |
| Asp462A | metal ligand |
| Arg339A | electrostatic stabiliser |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition
Step 3. The oxyanion collapses with the leaving CoA group being protonated from Arg339
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp462A | metal ligand |
| Glu434A | metal ligand |
| Arg339A | proton donor |
Chemical Components
ingold: bimolecular elimination, proton transfer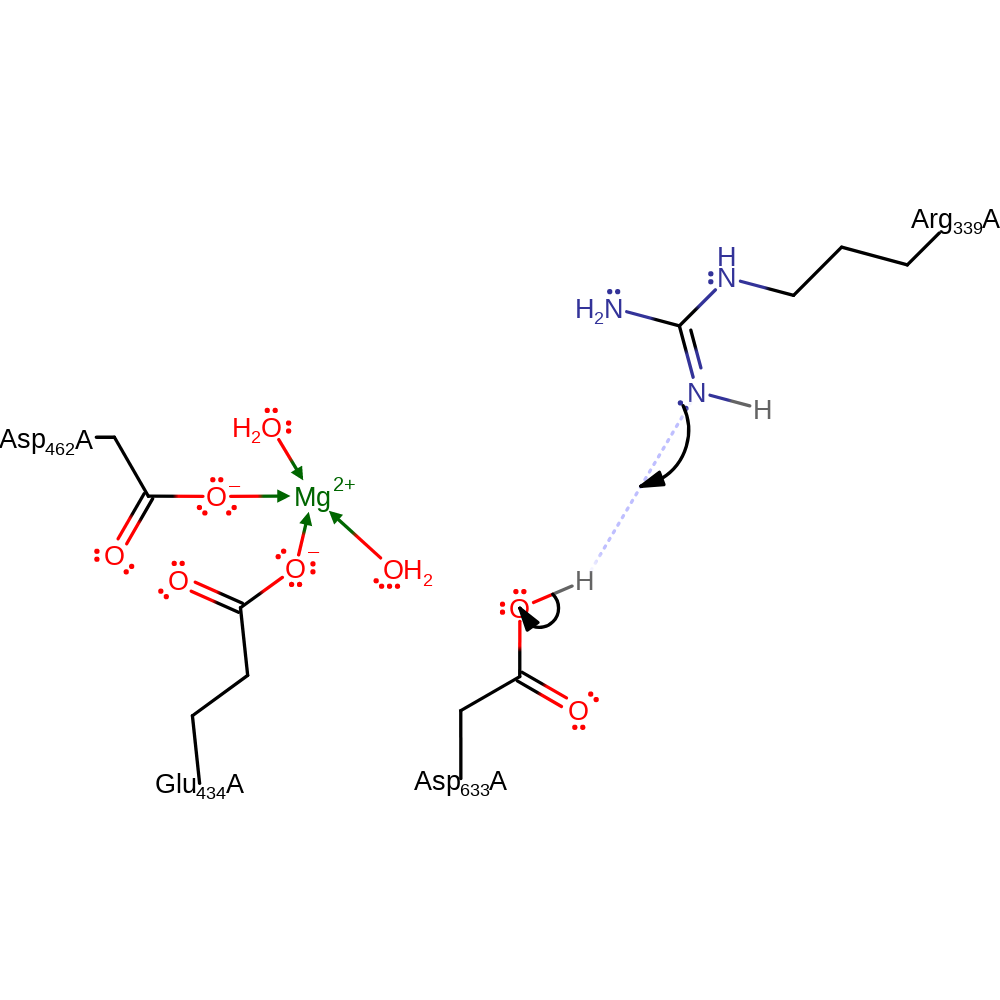
Step 4. Arg339 abstracts a proton from Asp633 in an inferred return step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu434A | metal ligand |
| Asp462A | metal ligand |
| Asp633A | proton donor |
| Arg339A | proton acceptor |






 Download:
Download: 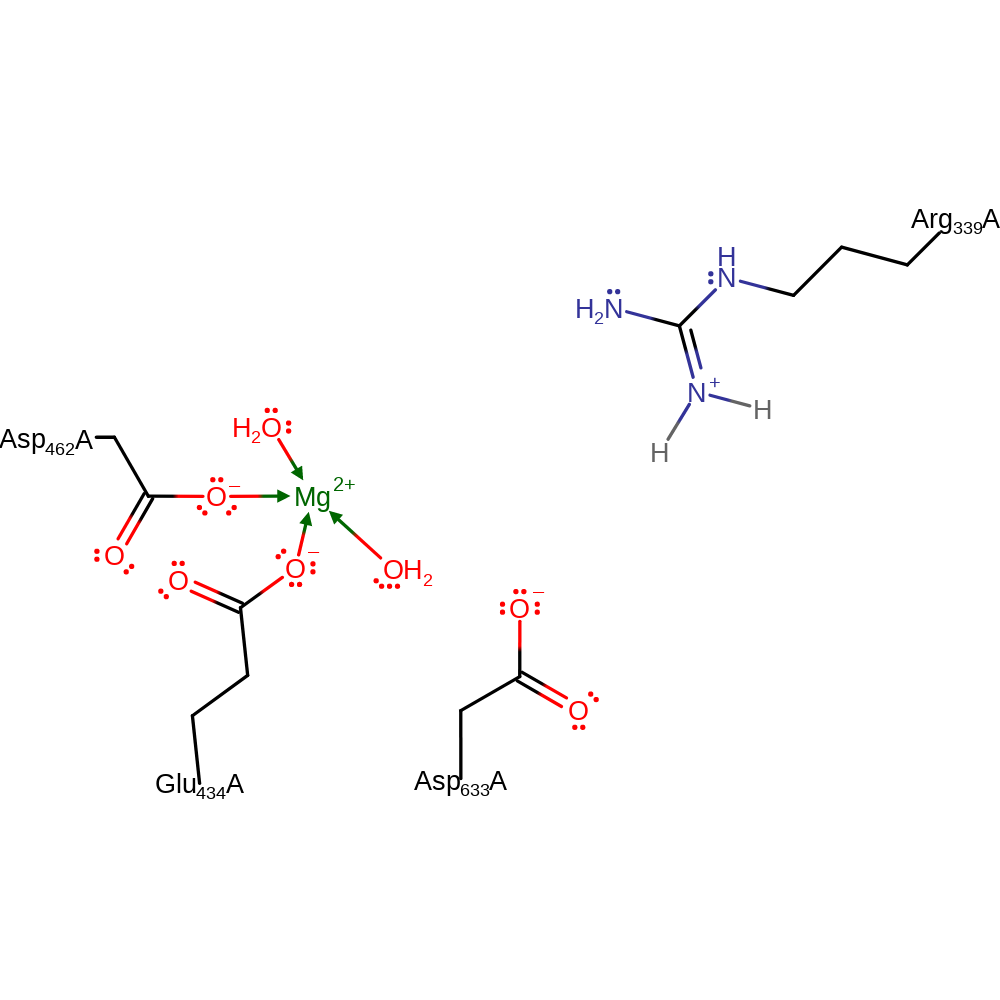 Download:
Download: