GTP cyclohydrolase I
GTP cyclohydrolase I catalyses the complex reaction which converts GTP to dihydroneopterin triphosphate. This is the first step of a pathway, which in plants and micro-organisms leads to tetrahydrofolate production, and in animals leads to tetrahydrobiopterin production. Tetrahydrobiopterin is biologically important as it serves as a cofactor in the production of catecholamines and nitric oxide. Genetic defects in GTP cyclohydrolase can therefore lead to severe neurological disorders. Enzymes involved in the formation of tetrahydrofolate are also important anti-infection drug targets.
Allosteric enzyme. Activity is modulated by K+, divalent cations, UTP, and tetrahydrobiopterin. Tetrahydrobiopterin is an inhibitor of this enzyme.
Reference Protein and Structure
- Sequence
-
P0A6T5
 (3.5.4.16)
(3.5.4.16)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1fbx
- CRYSTAL STRUCTURE OF ZINC-CONTAINING E.COLI GTP CYCLOHYDROLASE I
(2.8 Å)



- Catalytic CATH Domains
-
3.30.1130.10
 (see all for 1fbx)
(see all for 1fbx)
- Cofactors
- Zinc(2+) (1), Water (4) Metal MACiE
Enzyme Reaction (EC:3.5.4.16)
Enzyme Mechanism
Introduction
GTP cyclohydrolase I contains an essential zinc cation, thought to act as a Lewis acid, activating a water molecule towards hydrolytic opening of the imidazole ring of GTP. Three residues, Cys110, Cys181, His112 coordinate to this metal, with one vacant coordination position available to the hydrolytic water.
The reaction is initiated by His179, which acts as an acid and donates a proton to N7. The positively charged ring is now susceptible to nucleophilic attack at C8 by a water molecule. His112 protonates the bridging O of the furanose ring which leads to the opening of the furanose ring. Amadori rearrangement of this intermediate and proton abstraction by a base thought to be Ser135 leads to another intermediate. The last stage of the reaction, the closure of the pterin ring system is thought to be non-enzymatically catalysed either on the protein surface, or in solution.
Catalytic Residues Roles
| UniProt | PDB* (1fbx) | ||
| His180 | His179A | Acts as a general acid/base via an active site water molecule. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu112 | Glu111A | Activates His112 via a water molecule. | hydrogen bond acceptor, electrostatic stabiliser |
| His113 | His112A | Acts as a general acid base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Gln152 | Gln151A | Activates His179. | hydrogen bond acceptor, electrostatic stabiliser |
| His114, Cys111, Cys182 | His113A, Cys110A, Cys181A | Form the Zn(II) binding site. | metal ligand |
Chemical Components
aromatic bimolecular nucleophilic addition, overall reactant used, intermediate formation, proton transfer, bimolecular elimination, decyclisation, intramolecular nucleophilic addition, cyclisation, bimolecular nucleophilic substitution, overall product formed, unimolecular elimination by the conjugate base, rate-determining step, intramolecular rearrangement, keto-enol tautomerisation, intramolecular eliminationReferences
- Rebelo J et al. (2003), J Mol Biol, 326, 503-516. Biosynthesis of Pteridines. Reaction Mechanism of GTP Cyclohydrolase I. DOI:10.1016/s0022-2836(02)01303-7. PMID:12559918.
- Gräwert T et al. (2013), IUBMB Life, 65, 310-322. Structures and reaction mechanisms of GTP cyclohydrolases. DOI:10.1002/iub.1153. PMID:23457054.
- Ren J et al. (2005), J Biol Chem, 280, 36912-36919. GTP Cyclohydrolase II Structure and Mechanism. DOI:10.1074/jbc.m507725200. PMID:16115872.
- Tanaka Y et al. (2005), J Biochem, 138, 263-275. Novel Reaction Mechanism of GTP Cyclohydrolase I. High-Resolution X-Ray Crystallography of Thermus thermophilus HB8 Enzyme Complexed with a Transition State Analogue, the 8-Oxoguanine Derivative. DOI:10.1093/jb/mvi120. PMID:16169877.
- Lee S et al. (2002), BMB Rep, 35, 255-261. Biochemical Characterization of Oligomerization of Escherichia coli GTP Cyclohydrolase I. DOI:10.5483/bmbrep.2002.35.3.255.
- Schramek N et al. (2001), J Biol Chem, 276, 2622-2626. Ring Opening Is Not Rate-limiting in the GTP Cyclohydrolase I Reaction. DOI:10.1074/jbc.m004912200. PMID:11056154.
- Auerbach G et al. (2000), Proc Natl Acad Sci U S A, 97, 13567-13572. Zinc plays a key role in human and bacterial GTP cyclohydrolase I. DOI:10.1073/pnas.240463497. PMID:11087827.
- Bracher A et al. (1998), J Biol Chem, 273, 28132-28141. Biosynthesis of Pteridines: NMR STUDIES ON THE REACTION MECHANISMS OF GTP CYCLOHYDROLASE I, PYRUVOYLTETRAHYDROPTERIN SYNTHASE, AND SEPIAPTERIN REDUCTASE. DOI:10.1074/jbc.273.43.28132. PMID:9774432.
- Nar H et al. (1995), Structure, 3, 459-466. Atomic structure of GTP cyclohydrolase I. DOI:10.1016/s0969-2126(01)00179-4. PMID:7663943.
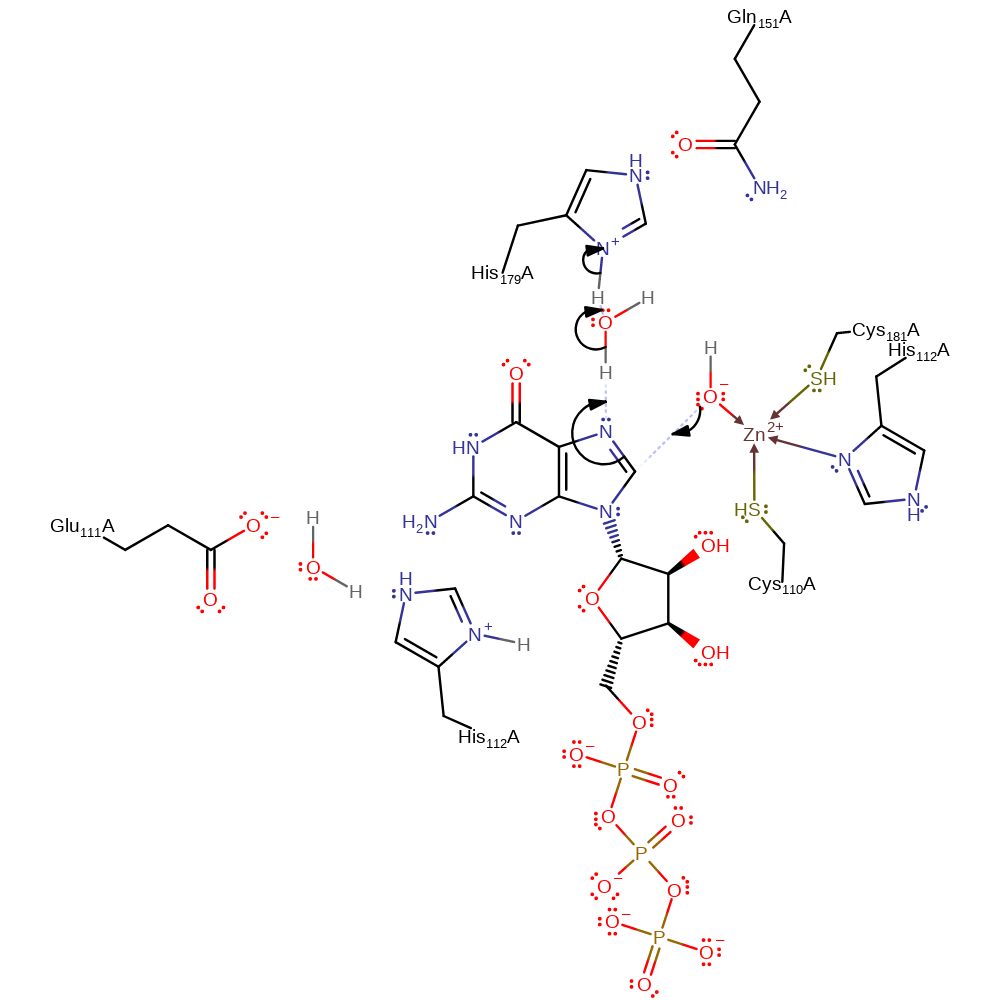
Step 1. Zinc activated water initiates a nucleophilic attack on the C8 of the substrate in an aromatic addition reaction. The nitrogen, N7, which receives the lone pair of electrons gains a proton from His112.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond donor |
| His112A | hydrogen bond donor |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
| His179A | proton donor |
Chemical Components
ingold: aromatic bimolecular nucleophilic addition, overall reactant used, intermediate formation, proton transfer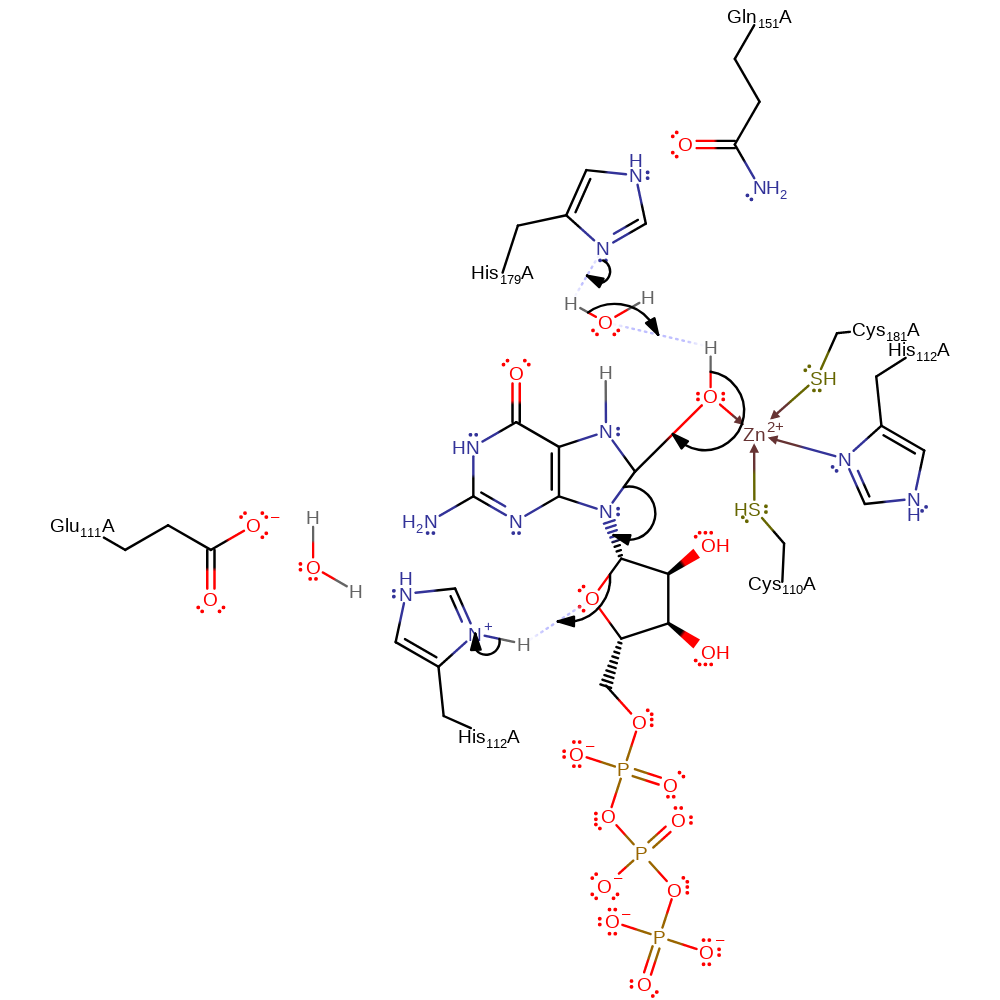
Step 2. His179 deprotonates the added hydroxyl group, cleaving the C8-N9 bond in an elimination which results in the cleavage of the C1-O4 bond in the ribose ring and concomitant deprotonation of His112 by O4.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond acceptor |
| His112A | hydrogen bond donor |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
| Gln151A | electrostatic stabiliser |
| Glu111A | electrostatic stabiliser, hydrogen bond acceptor |
| Gln151A | hydrogen bond acceptor |
| His179A | proton acceptor |
| His112A | proton donor |
Chemical Components
ingold: bimolecular elimination, decyclisation, intermediate formation, proton transfer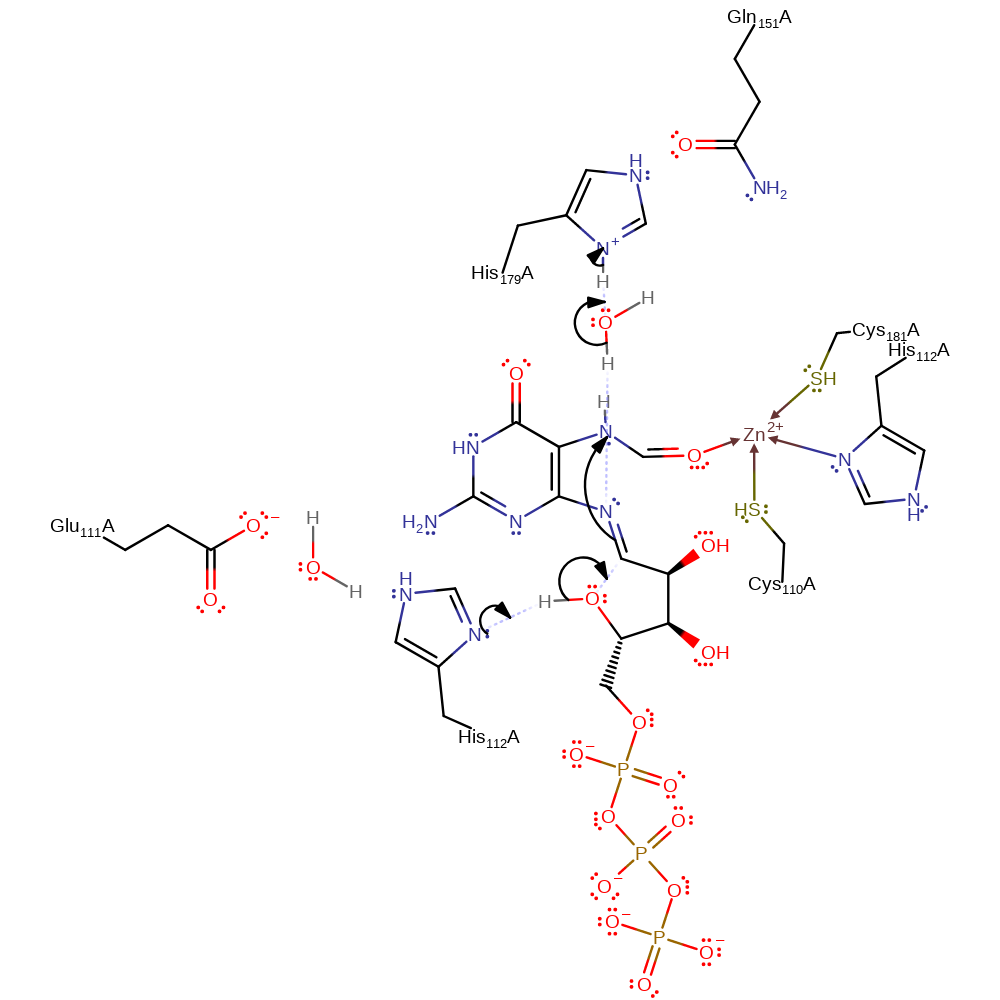
Step 3. His112 deprotonates the newly formed hydroxyl group, which initiates the reformation of the ribose ring in a nucleophilic addition reaction, which causes the N9 to deprotonate His179.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond donor |
| His112A | hydrogen bond acceptor |
| Glu111A | hydrogen bond acceptor |
| Gln151A | hydrogen bond acceptor |
| Glu111A | electrostatic stabiliser |
| Gln151A | electrostatic stabiliser |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
| His179A | proton donor |
| His112A | proton acceptor |
Chemical Components
ingold: intramolecular nucleophilic addition, cyclisation, intermediate formation, proton transfer
Step 4. His179 acts as a general base, deprotonating water, which is activated by zinc, this hydroxide then initiates a nucleophilic attack on the amide carbon of the intermediate in a substitution reaction which eliminates formate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond acceptor |
| His112A | hydrogen bond donor |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
| His179A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation, proton transfer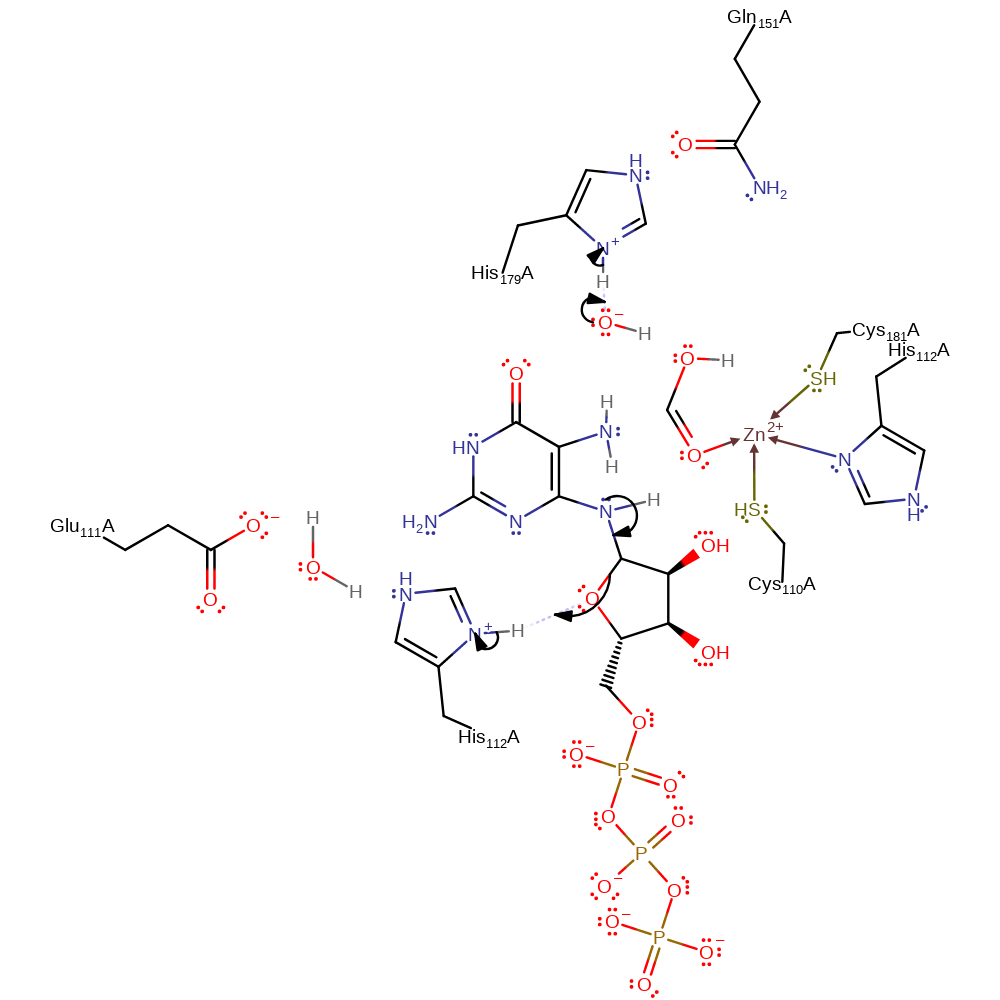
Step 5. First step in the Amadori Rearrangement. His112 donates a proton to the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond acceptor |
| His112A | hydrogen bond donor |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
| His112A | proton donor |
| His179A | proton donor |
Chemical Components
intermediate formation, proton transfer
Step 6. Second step of the Amadori Rearrangement . Tautomerisation of the N=C-C bonds.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond acceptor |
| His112A | hydrogen bond acceptor |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
Chemical Components
ingold: unimolecular elimination by the conjugate base, intermediate formation, rate-determining step
Step 7. Next step of the Amadori Rearrangement. Keto-enol tautomerisation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond acceptor |
| His112A | hydrogen bond acceptor |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
Chemical Components
proton transfer, intramolecular rearrangement, intermediate formation, keto-enol tautomerisation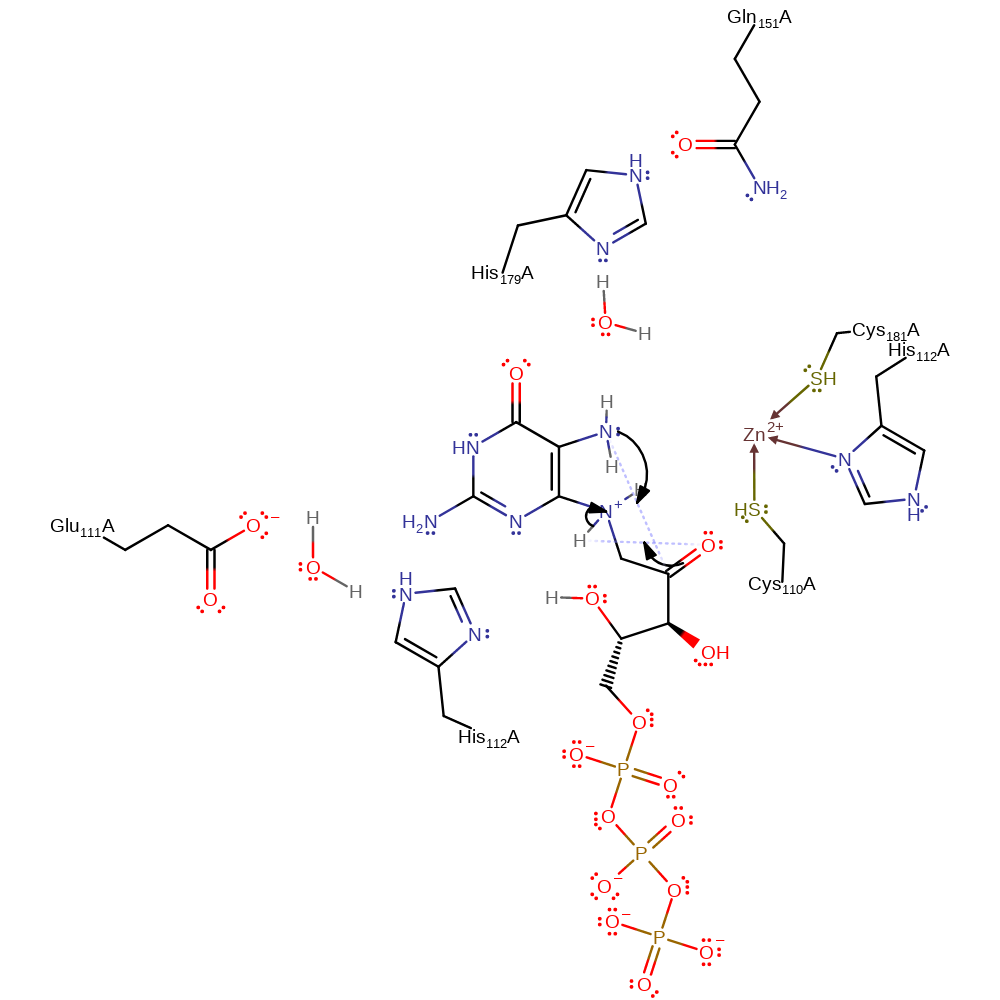
Step 8. Final step of the Amadori Rearrangement. N7 initiates a nucleophilic attack on the carbonyl carbon, forming the new six-membered ring.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond acceptor |
| His112A | hydrogen bond acceptor |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
Chemical Components
proton transfer, intermediate formation, cyclisation, ingold: intramolecular nucleophilic addition
Step 9. Water is eliminated, forming a new double bond which extends the conjugation across the molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond donor |
| His112A | hydrogen bond acceptor |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
Chemical Components
intermediate formation, proton transfer, ingold: intramolecular elimination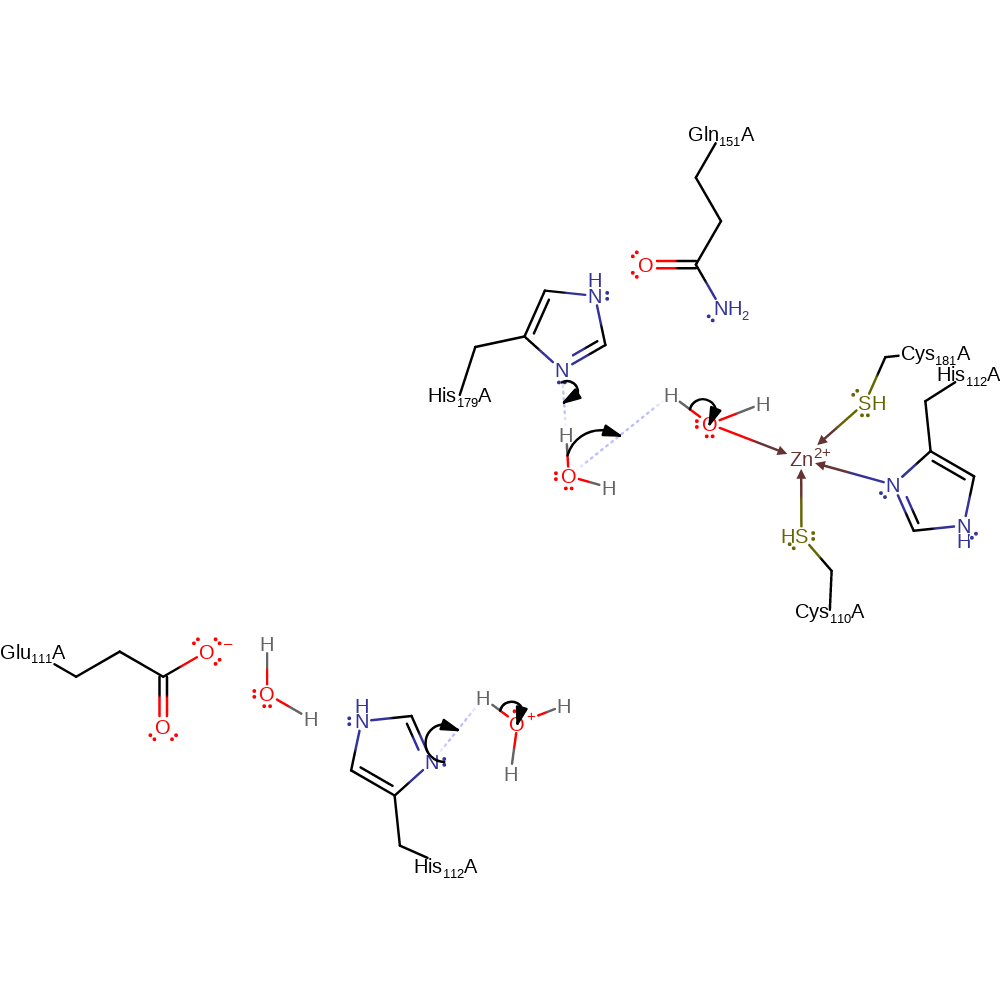
Step 10. Inferred return step in which His179 and 112 are reprotonated from bulk solvent water molecules, and the activated water molecule zinc ligand is regenerated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His179A | hydrogen bond donor |
| His112A | hydrogen bond acceptor |
| Cys181A | metal ligand |
| Cys110A | metal ligand |
| His113A | metal ligand |
| Glu111A | electrostatic stabiliser |
| Gln151A | electrostatic stabiliser |
| His179A | proton acceptor |
| His112A | proton acceptor |




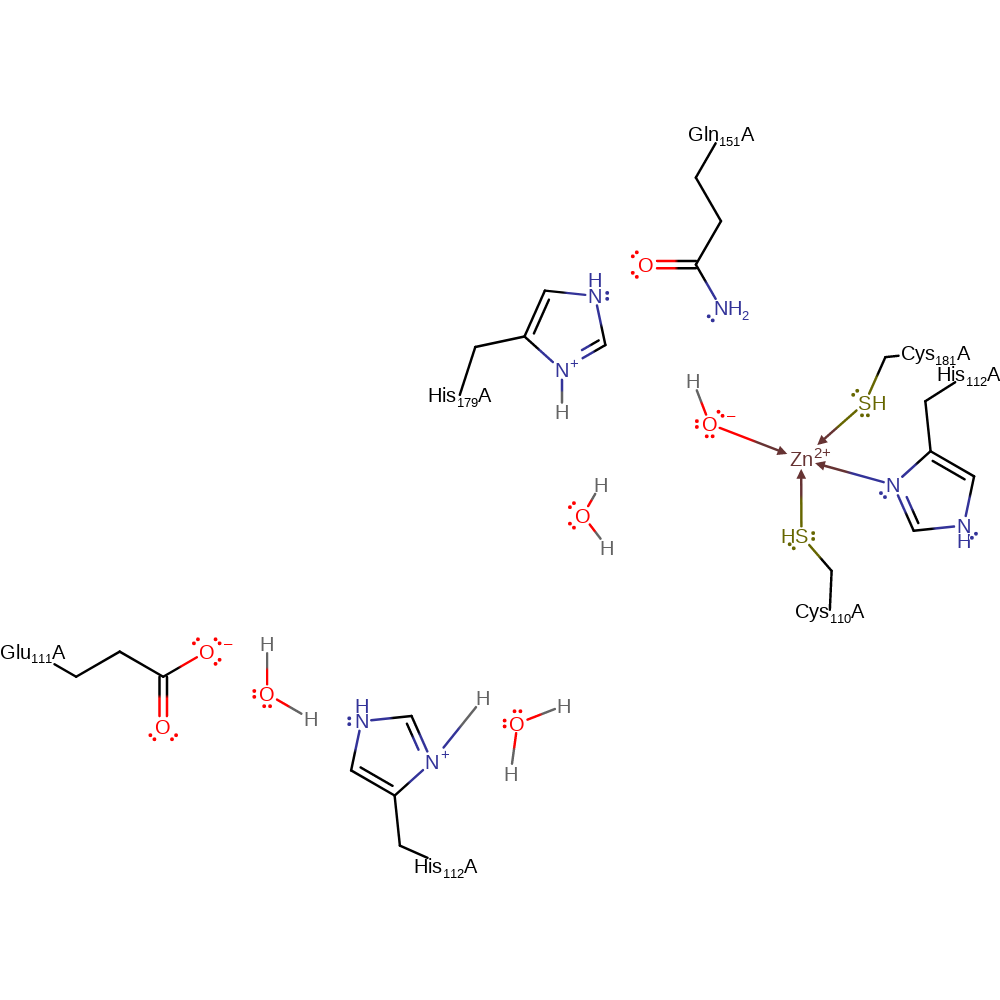 Download:
Download: