Glycine N-methyltransferase
Glycine N-methyltransferase (GNMT) isolated from Rattus norvegicus catalyses the transfer of a methyl group from S-adenosyl-methionine (SAM) to glycine to produce N-methylglycine (sarcosine) and S-adenosyl homocysteine (SAH). Unlike most SAM-dependent methyltransferases, GNMT is not strongly inhibited by SAH. This, coupled with no evidence of a physiological role for sarcosine, has led to the suggestion that GNMT plays a key role in modulating the SAM/SAH ratio in tissues where it is an important cofactor, and so controls methyltransferase activity.
GNMT exists as a dimer of dimers. Each subunit contains a molecular basket structure and a flexible N-terminal U-loop that can block the entrance to the basket of the partner subunit of the dimer. The U-loop competes with SAM for binding at the active site and so can regulate catalytic activity. GNMT binds first to SAM, which causes a conformational change, and then to glycine.
Reference Protein and Structure
- Sequence
-
P13255
 (2.1.1.20)
(2.1.1.20)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rattus norvegicus (Norway rat)

- PDB
-
1xva
- METHYLTRANSFERASE
(2.2 Å)



- Catalytic CATH Domains
-
3.30.46.10
 3.40.50.150
3.40.50.150  (see all for 1xva)
(see all for 1xva)
- Cofactors
- S-adenosyl-l-methionine (1)
Enzyme Reaction (EC:2.1.1.20)
Enzyme Mechanism
Introduction
SAM and glycine bind in the active site in sequential order (SAM first). Glycine binds to the NE of tryptophan and the main chain carbonyl of alanine. Hydrogen bond interactions between glutamate and the SAM and glutamate and the glycine encourage the nucleophilic attack by glycine on the SAM methyl group. However, GNMT primarily catalyses the reaction through proximity and orientation effects. Arg175 favours the binding of the basic form of glycine (amine group neutral and carboxylate group deprotonated) over the zwitterionic form. Glycine and SAM are bound in such a way to align the lone pair orbital of the amino nitrogen of the amine and C-S bond of the methyl group of SAM. Tyr21 is thought to polarise this bond, though there is conflicting evidence on the subject. The thermal motion of the enzyme causes the two substrates to collide, leading to an Sn2 reaction to form sarcosine and SAH. The transition state of this reaction is stabilised by hydrogen bonds between the amine group of glycine and Gly137 and Tyr242. It is better stabilised when glycine is in the basic form than the zwitterionic form.
Catalytic Residues Roles
| UniProt | PDB* (1xva) | ||
| His143 | His142A | His142 accepts a hydrogen bond from the hydroxyl group of Tyr21, freeing up the lone pair of Tyr21 to stabilise the S+ of SAM | activator |
| Tyr22 | Tyr21A | Tyr21 forms an electrostatic interaction with the S+ of SAM, stabilising the transition state. | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Tyr195 | Tyr194A | Tyr194 forms a hydrogen bond to the amine group of glycine. This bond strengthens upon moving to the transition state, thus stabilising it | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Gly138 (main-C) | Gly137A (main-C) | The carbonyl group of main chain Gly137 forms a hydrogen bond to the amine group of glycine. This bond strengthens upon moving to the transition state, thus stabilising it. | hydrogen bond acceptor, electrostatic stabiliser |
| Arg176 | Arg175A | Arg175 is selective for the basic form of glycine (amine group neutral and carboxylic group deprotonated) over the zwitterionic tautomer. This provides better stabilisation of the transition state. | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall product formed, overall reactant used, native state of cofactor is not regenerated, native state of enzyme regeneratedReferences
- Soriano A et al. (2006), Biochemistry, 45, 14917-14925. Catalysis in GlycineN-Methyltransferase: Testing the Electrostatic Stabilization and Compression Hypothesis†. DOI:10.1021/bi061319k. PMID:17154529.
- Velichkova P et al. (2005), J Phys Chem B, 109, 8216-8219. Methyl Transfer in GlycineN-Methyltransferase. A Theoretical Study. DOI:10.1021/jp0443254. PMID:16851960.
- Takata Y et al. (2003), Biochemistry, 42, 8394-8402. Catalytic Mechanism of GlycineN-Methyltransferase†,∇. DOI:10.1021/bi034245a. PMID:12859184.
- Fu Z et al. (1996), Biochemistry, 35, 11985-11993. Crystal Structure of GlycineN-Methyltransferase from Rat Liver†,‡. DOI:10.1021/bi961068n. PMID:8810903.
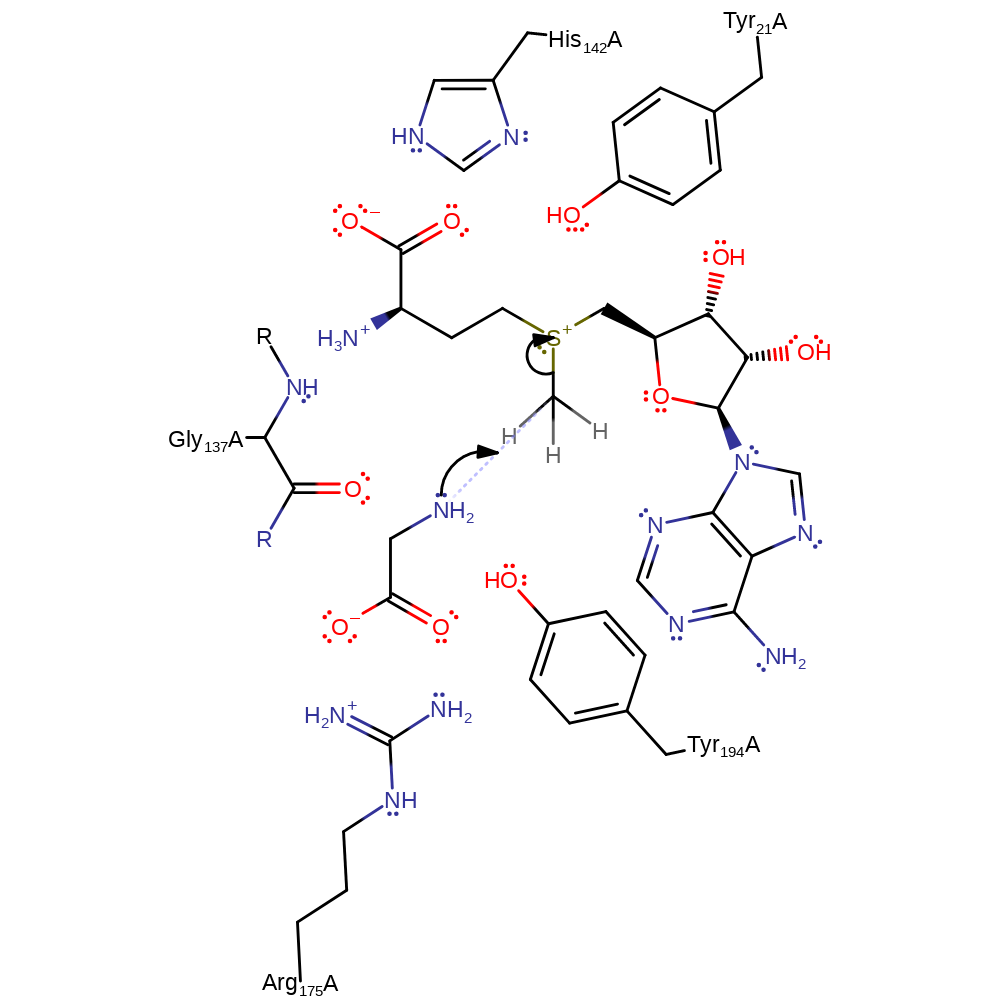
Step 1. The amine of the substrate glycine initiates a nucleophilic attack on the methyl of the S-adenosyl-L-methionine in a substitution reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly137A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Tyr21A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Tyr194A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| His142A | activator |
| Arg175A | electrostatic stabiliser |





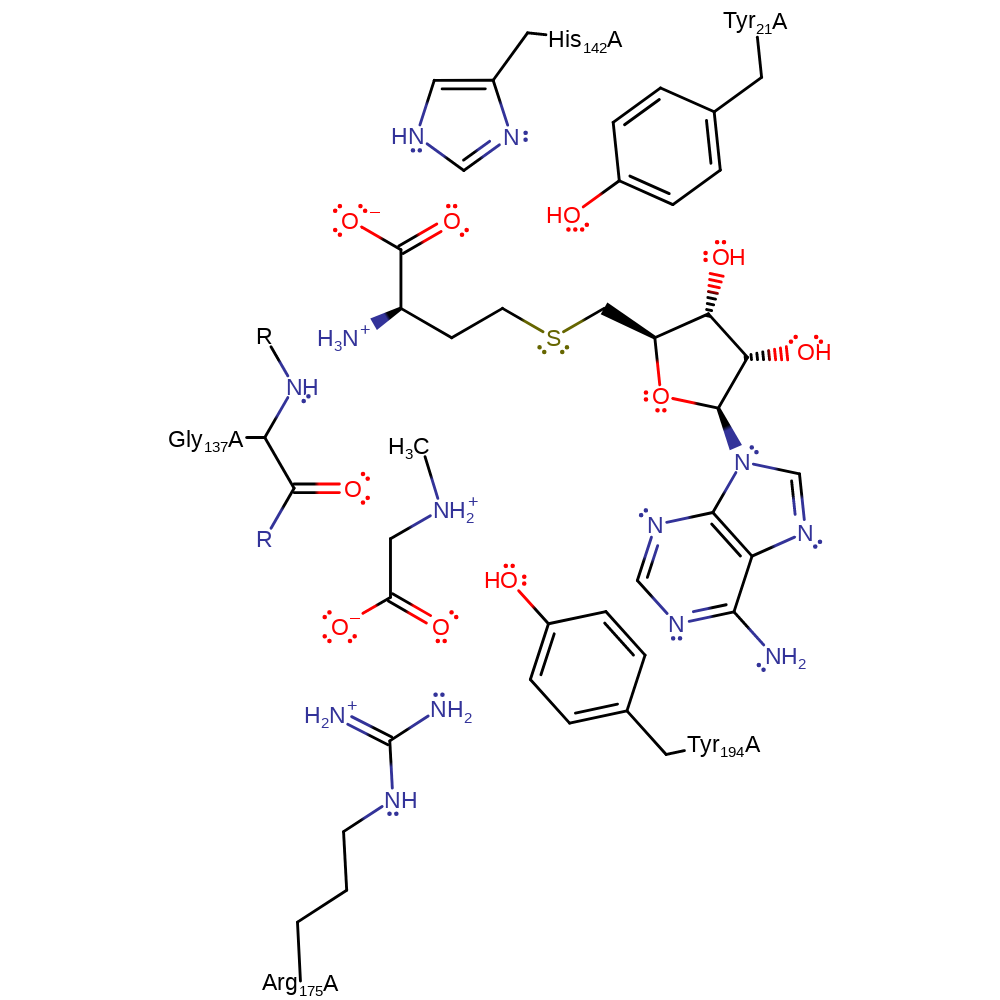 Download:
Download: