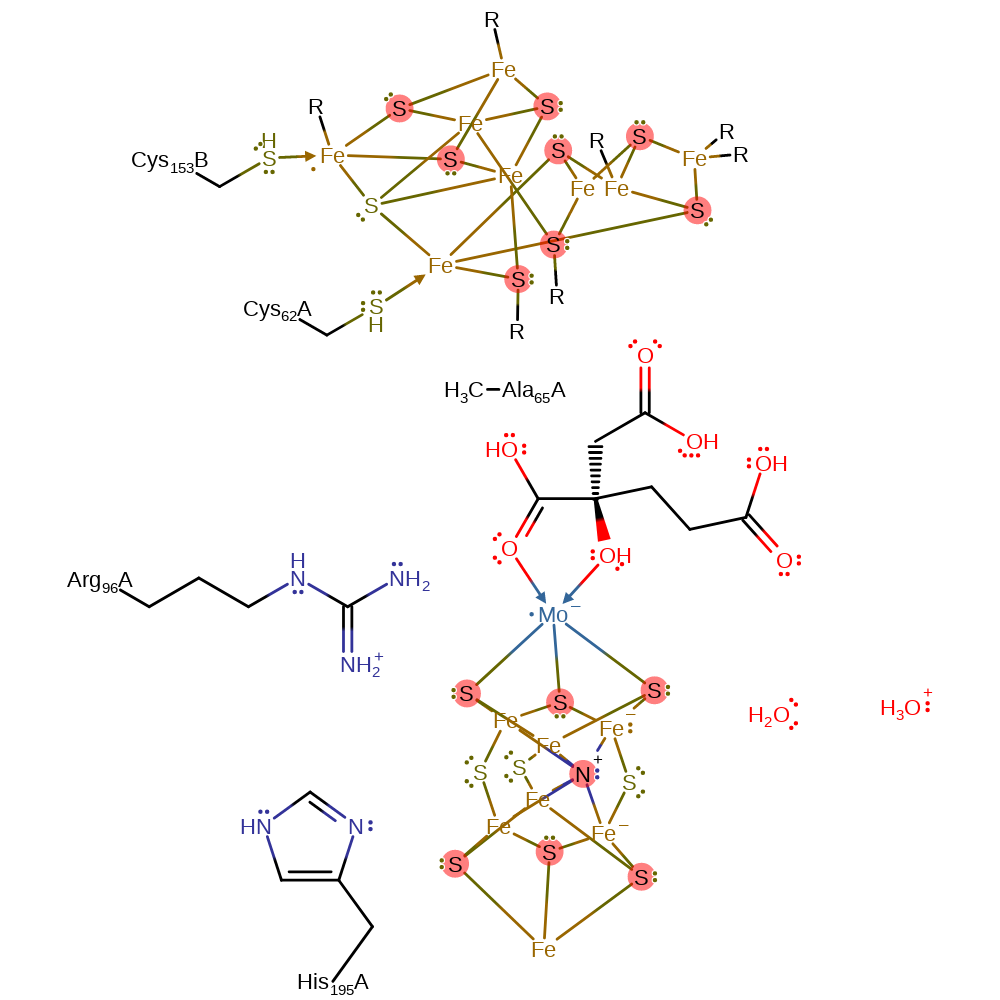
Nitrogenase
The nitrogenase complex from Azotobacter vinelandii catalyses the reduction of atmospheric nitrogen to ammonia, coupled to the hydrolysis of ATP and the oxidation of ferrodoxin. It is composed of two enzyme components: the Fe protein and the MoFe protein. The Fe protein catalyses the hydrolysis of ATP and consequently transfers electron to the MoFe protein. The MoFe protein contains the FeMo-cofactor which is composed of molybdenum, a [7Fe-9S] cluster, and homocitrate, and the P-cluster which is composed of a [8Fe-7S] cluster. The reducing electrons ultimately come from an external electron donor such as ferredoxin, through the Fe protein to the FeMo-cofactor [PMID:17614310, PMID:14551236]. The complex is formed as a heterotetramer of the FeMo protein, with one homodimer of Fe protein associated with each of the two alpha-beta subunits. The FeMo protein contains the FeMo-cofactor which contains a Fe7MoS9-homocitrate cofactor (FeMo-co) which is thought to contain a nitrogen atom, and the P-cluster containing an [8Fe-7S] cluster. The Fe protein contains a [4Fe-4S] cubane cluster.
Nitrogenase is very biologically important because it reduces inert dinitrogen into a form which organisms can use in synthesis. Only a small subset of organisms can catalyse this reaction while all organisms require reduced nitrogen. This reaction is of industrial interest because enzymatic nitrogen fixation occurs under very mild conditions, while the industrial Haber-Bosch process must be run at high temperatures and pressures.
It should be noted that the mechanism of this enzyme is still very unclear and that the exact atoms in the FeMo-cofactor involved are unknown. However, it is known that the Fe2, S2B, Fe6, S3b, Fe7, S5A, Fe3 and S2A make up the reactive face of the cofactor [PMID:17614310].
Reference Protein and Structure
- Sequences
-
P07328
 (1.18.6.1)
(1.18.6.1)
P07329 (1.18.6.1)
(1.18.6.1)
P00459 (1.18.6.1)
(1.18.6.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Azotobacter vinelandii (Bacteria)

- PDB
-
1n2c
- NITROGENASE COMPLEX FROM AZOTOBACTER VINELANDII STABILIZED BY ADP-TETRAFLUOROALUMINATE
(3.0 Å)



- Catalytic CATH Domains
-
3.40.50.1980
 3.40.50.300
3.40.50.300  (see all for 1n2c)
(see all for 1n2c)
- Cofactors
- Fe8s7 iron-sulfur cluster (1), Tetra-mu3-sulfido-tetrairon (1), Magnesium(2+) (1), Iron-sulfur-molybdenum cluster (1), (2r)-homocitric acid (1) Metal MACiE
Enzyme Reaction (EC:1.18.6.1)
Enzyme Mechanism
- Summary
- Step 1
- Step 2
- Step 3
- Step 4
- Step 5
- Step 6
- Step 7
- Step 8
- Step 9
- Step 10
- Step 11
- Step 12
- Step 13
- Step 14
- Step 15
- Products
- All Steps
Introduction
The Fe protein and the FeMo protein must associate before catalytic reactions can occur. Once associated, Asp129 from the Fe protein removes a proton from water and hydroxide acts as the nucleophile in the hydrolysis of ATP to ADP and phosphate. This causes a conformational change that enables the transfer of an electron from the [4Fe-4S] cluster of the Fe protein to the P-cluster and then the FeMo-co of the FeMo protein. The protein complex then dissociates and the Fe protein is reduced by an external molecule of reduced ferrodoxin. For each molecule of ammonia produced, this cycle of ATP hydrolysis and electron transfer to the FeMo protein must occur eight times. The pathway of electron transfer to the FeMo-co is poorly understood, though the electron is thought to pass through the homocitrate ligand before being transferred to the active face of the cluster. This allows S3B to be protonated, with the proton source thought to be a water chain. The proton is then transferred to a different atom in the complex, thought to be iron, and S3B accepts another proton. Dihydrogen is then released and two more protons are transferred to the cluster from the bulk solvent. Dinitrogen can then bind to an iron in an end-on fashion, and then bind to another iron in a similar fashion. There is proton transfer to one and then the other of the nitrogens. This is followed by an intramolecular nucleophilic substitution, leaving a nitrogen bridging the two irons and bonded to an NH3+ group. There is then N-N bond cleavage (mechanism unclear) to release one molecule of ammonia and to leave the remaining nitrogen bonded to two irons and two protons. A substitution reaction occurs that leads to the cleavage of one Fe-N bond, followed by proton transfer to the nitrogen. There is then a second nucleophilic substitution to release the second molecule of ammonia.
Catalytic Residues Roles
| UniProt | PDB* (1n2c) | ||
| Asp130 | Asp129E | Acts as a general base for water attack on the terminal phosphate group of ATP. It also stabilises the terminal phosphate group during the reaction. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Val157 (main), Cys153 | Val157(156)B (main), Cys153(152)B | Acts as an electron donor/acceptor in the transfer of an electron from the Fe protein to the P-cluster of the FeMo protein. | single electron relay, single electron acceptor, single electron donor, polar interaction |
| Lys11, Lys42, Lys16 | Lys10E, Lys41F, Lys15F | Stabilises the terminal phosphate group during ATP hydrolysis. | hydrogen bond donor, electrostatic stabiliser |
| Cys62, Ala65 | Cys62(61)A, Ala65(64)A | Forms part of the electron transfer pathway between the P-cluster and the FeMo-co. | metal ligand |
| His195, Arg96 | His195(194)A, Arg96(95)A | Forms a hydrogen bond with S2B of the FeMo-co during the resting state and acts as an activator. | activator, polar interaction |
Chemical Components
bimolecular nucleophilic substitution, electron transfer, proton transfer, dephosphorylation, hydrolysis, intermediate formation, electron relay, proton relay, overall product formed, cofactor used, native state of cofactor regenerated, overall reactant used, intermediate terminated, native state of enzyme regenerated, hydride transfer, intramolecular nucleophilic addition, intramolecular elimination, bimolecular nucleophilic addition, intramolecular electrophilic addition, intramolecular nucleophilic substitution, elimination (not covered by the Ingold mechanisms)References
- Dance I (2007), Chem Asian J, 2, 936-946. Elucidating the Coordination Chemistry and Mechanism of Biological Nitrogen Fixation. DOI:10.1002/asia.200700131. PMID:17614310.
- Rao L et al. (2016), ACS Catal, 6, 1567-1577. Theoretical Investigation on the Role of the Central Carbon Atom and Close Protein Environment on the Nitrogen Reduction in Mo Nitrogenase. DOI:10.1021/acscatal.5b02577.
- Varley JB et al. (2015), Phys Chem Chem Phys, 17, 29541-29547. Mechanistic insights into nitrogen fixation by nitrogenase enzymes. DOI:10.1039/c5cp04034e. PMID:26366854.
- Dance I (2008), Dalton Trans, 5977-5991. The chemical mechanism of nitrogenase: calculated details of the intramolecular mechanism for hydrogenation of eta(2)-N(2) on FeMo-co to NH(3). DOI:10.1039/b806100a. PMID:19082054.
- Kästner J et al. (2007), J Am Chem Soc, 129, 2998-3006. Ammonia Production at the FeMo Cofactor of Nitrogenase: Results from Density Functional Theory. DOI:10.1021/ja068618h. PMID:17309262.
- Tezcan FA et al. (2005), Science, 309, 1377-1380. Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein. DOI:10.1126/science.1115653. PMID:16123301.
- Igarashi RY et al. (2003), Crit Rev Biochem Mol Biol, 38, 351-384. Nitrogen Fixation: The Mechanism of the Mo-Dependent Nitrogenase. DOI:10.1080/10409230391036766. PMID:14551236.
- Torres RA et al. (2003), J Am Chem Soc, 125, 1923-1936. Density Functional and Reduction Potential Calculations of Fe4S4Clusters. DOI:10.1021/ja0211104. PMID:12580620.
- Chiu H et al. (2001), Biochemistry, 40, 641-650. MgATP-Bound and Nucleotide-Free Structures of a Nitrogenase Protein Complex between the Leu 127Δ-Fe-Protein and the MoFe-Protein†,‡. DOI:10.1021/bi001645e. PMID:11170380.
- Schindelin H et al. (1997), Nature, 387, 370-376. Structure of ADP·AIF4 –-stabilized nitrogenase complex and its implications for signal transduction. DOI:10.1038/387370a0. PMID:9163420.
- Burgess BK et al. (1996), Chem Rev, 96, 2983-3012. Mechanism of Molybdenum Nitrogenase. DOI:10.1021/cr950055x. PMID:11848849.
- Lanzilotta WN et al. (1995), Biochemistry, 34, 10713-10723. Nucleotide Hydrolysis and Protein Conformational Changes in Azotobacter vinelandii Nitrogenase Iron Protein: Defining the Function of Aspartate 129. DOI:10.1021/bi00034a003. PMID:7662655.

Step 1.
Hydrolysis of the MgATP (via Asp129E abstracting a proton from water) is thought to cause a conformational change in the protein which enables an electron to be transferred from the Fe4S4 cluster to the FeMo cofactors.
After hydrolysis and electron transfer the two proteins dissociate, allowing for the Fe protein to be re-reduced (step 2). Whilst this reaction is only shown once here, it is known that the cycle of steps 1 and 2 must occur eight times for the complete reduction of dinitrogen to ammonia (six times for the dinitrogen reduction, and a further two for the stoichiometric reduction of two protons that also occurs). The exact role of the P-cluster (part of the electron transfer pathway) is not entirely clear and is thought to be transient as no intermediate oxidation states have been observed, thus electron transfer from the P-cluster to the FeMo-cofactor is thought to occur very quickly, whilst electron transfer from the Fe4S2 cluster to the P-cluster is thought to be slow. The protonation of the FeMo-cofactor is thought to occur via a water chain that connects the cofactor with the surface of the enzyme (the entire chain is not shown here). The first atom to be protonated is the S3B, from which the protons can migrate through several pathways to various other S and Fe atoms within the cofactor .
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His195(194)A | polar interaction, activator |
| Lys41F | hydrogen bond donor, electrostatic stabiliser |
| Cys153(152)B | metal ligand |
| Lys15F | hydrogen bond donor, electrostatic stabiliser |
| Lys10E | hydrogen bond donor, electrostatic stabiliser |
| Val157(156)B (main) | polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| Asp129E | hydrogen bond acceptor |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Val157(156)B (main) | single electron acceptor |
| Asp129E | proton acceptor |
| Val157(156)B (main) | single electron relay |
| Ala65(64)A | single electron relay |
| Val157(156)B (main) | single electron donor |
| Ala65(64)A | single electron acceptor, single electron donor |
Chemical Components
ingold: bimolecular nucleophilic substitution, electron transfer, proton transfer, dephosphorylation, hydrolysis, intermediate formation, electron relay, proton relay, overall product formed, cofactor used, native state of cofactor regenerated, overall reactant used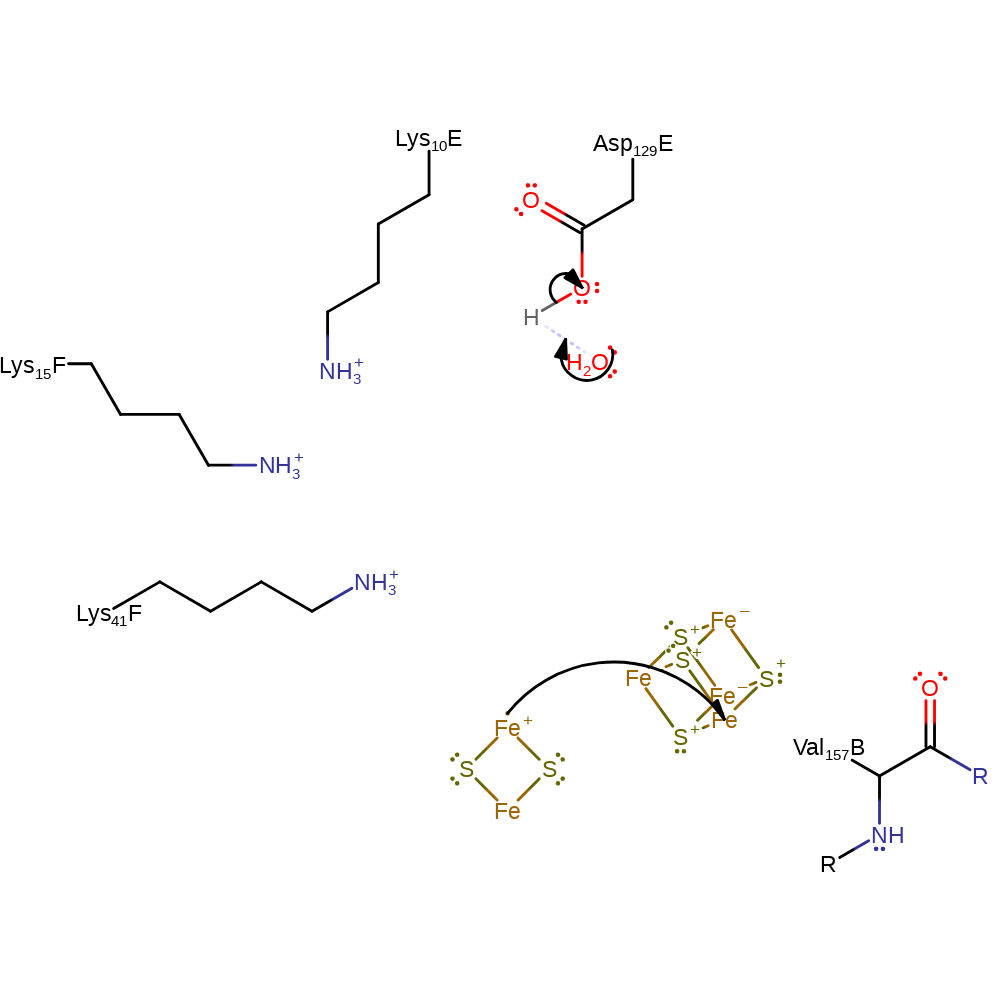
Step 2. Once the electron has been transferred from the Fe protein to the MoFe protein, the two proteins dissociate (thought to be rate limiting),. This step occurs only in the Fe protein, and involves the exchange of ATP back into the Fe protein and also the reduction of the Fe protein using an external reducing agent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp129E | hydrogen bond donor |
| Asp129E | proton donor |
Chemical Components
electron transfer, proton transfer, intermediate terminated, proton relay, native state of cofactor regenerated, overall reactant used, overall product formed, electron relay, native state of enzyme regenerated
Step 3. The proton on the S3B is transferred to a different atom in the cofactor and a second proton is transferred via the water chain.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
proton transfer, intermediate formation, proton relay, overall reactant used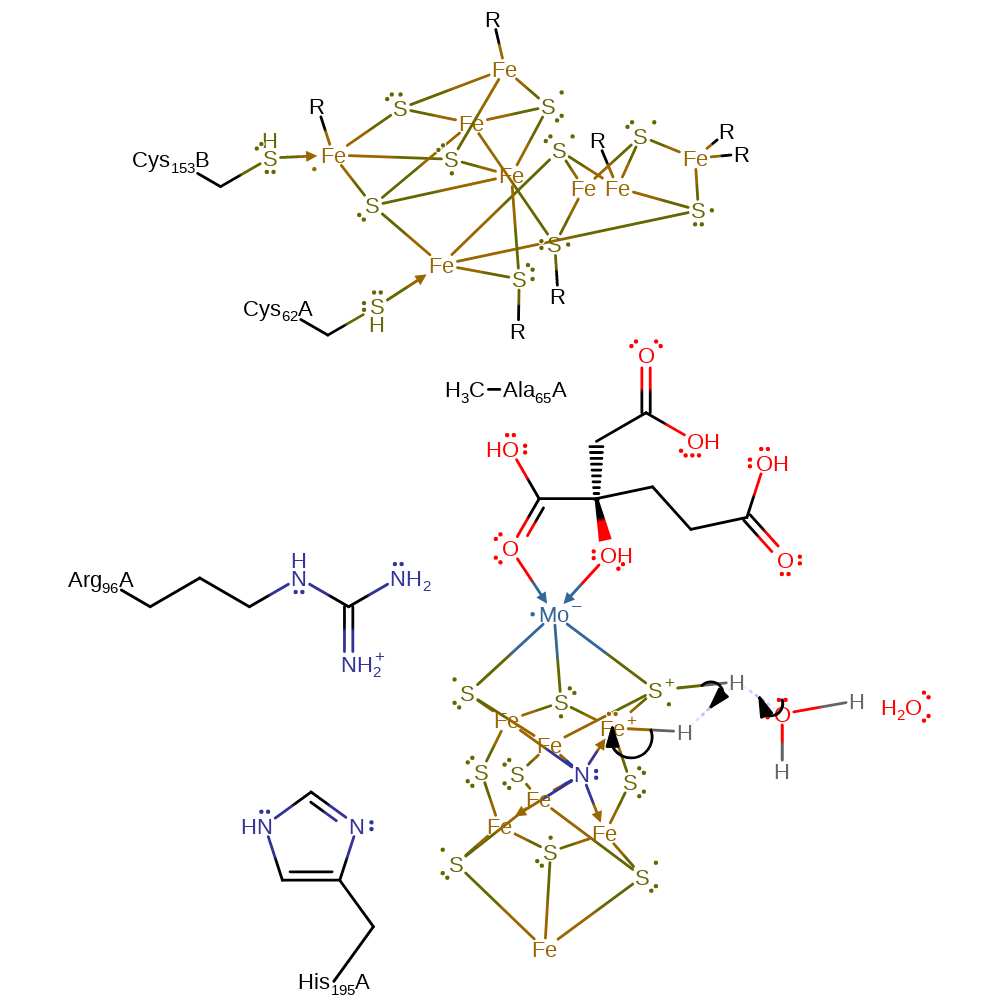
Step 4. Dihydrogen is eliminated from the Fe-Mo-S cluster cofactor. Formation of the dihydrogen is required before the dinitrogen can bind and requires the accumulation of at least two electron equivalents in the MoFe protein. The location of the dihydrogen molecule within the FeMo-cofactor is arbitary.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
proton transfer, hydride transfer, ingold: intramolecular nucleophilic addition, ingold: intramolecular elimination, intermediate formation, overall product formed
Step 5. Dinitrogen will only bind to the FeMo-cofactor when it reaches an appropriate oxidation state. Before dinitrogen is bound, a further two protons are transferred from the bulk solvent to the FeMo-cofactor. Dinitrogen is then bound to one of the iron atoms in the Fe-Mo-S cluster cofactor in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 6. Dinitrogen is bound in a bidentate manner through an electrophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
ingold: intramolecular electrophilic addition, intermediate formation
Step 7. A proton is transferred to the bound dinitrogen from the S3B thiol, which reprotonates from water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
proton transfer, intermediate formation, proton relay, overall reactant used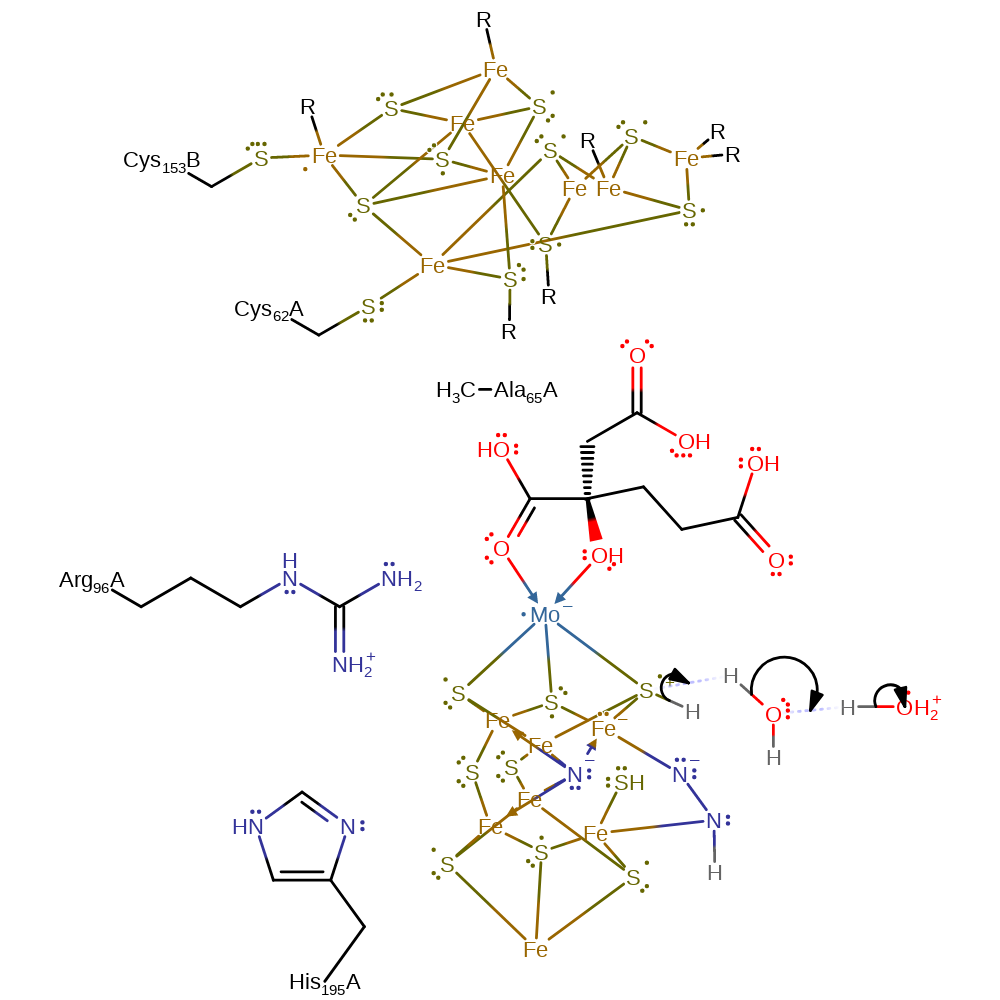
Step 8. A second proton is transferred to the bound dinitrogen from the S3B thiol, which reprotonates from water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
proton transfer, intermediate formation, proton relay, overall reactant used
Step 9. In a nucleophilic substitution, the diimide becomes bound in a monodentate manner, and another proton is transferred from the S3B thiol, which reprotonates from water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
ingold: intramolecular nucleophilic substitution, proton transfer, intermediate formation, overall reactant used, proton relay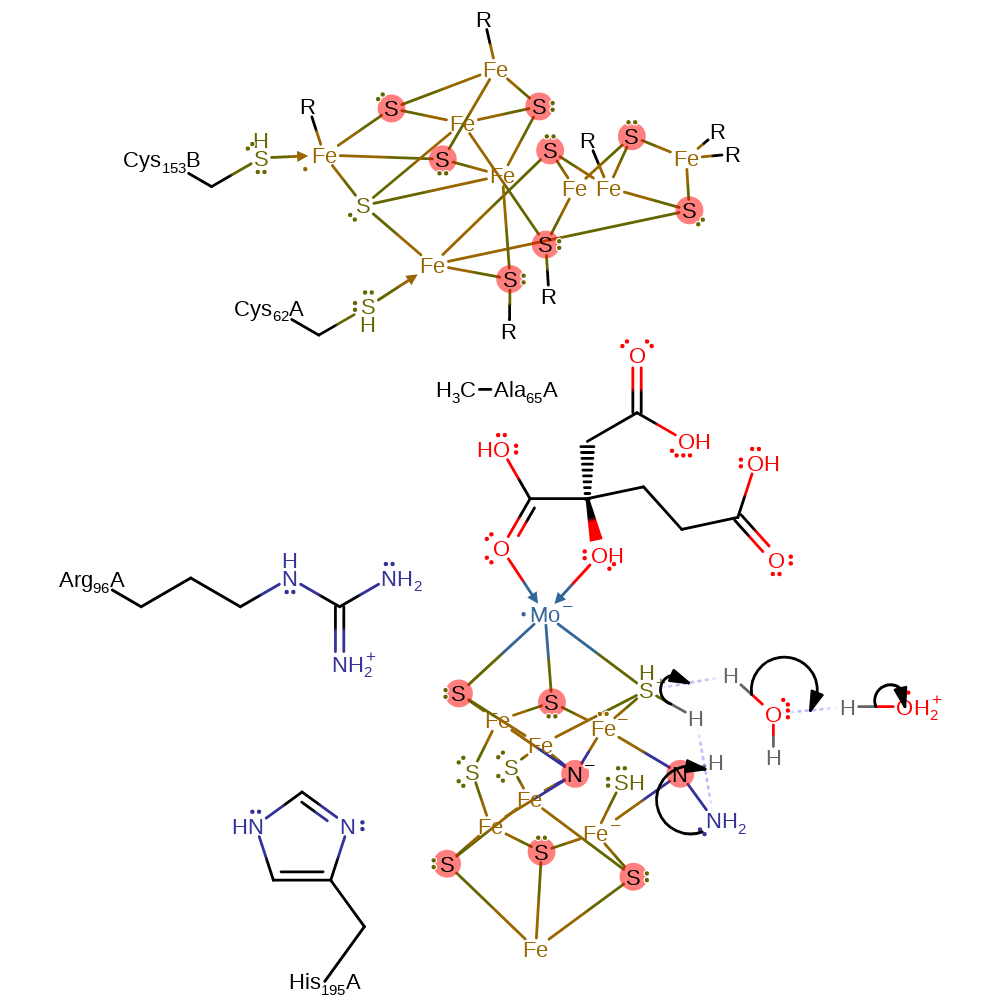
Step 10. The hydrazine intermediate gains another proton from the S3B thiol, which reprotonates from water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
proton transfer, intermediate formation, proton relay, overall reactant used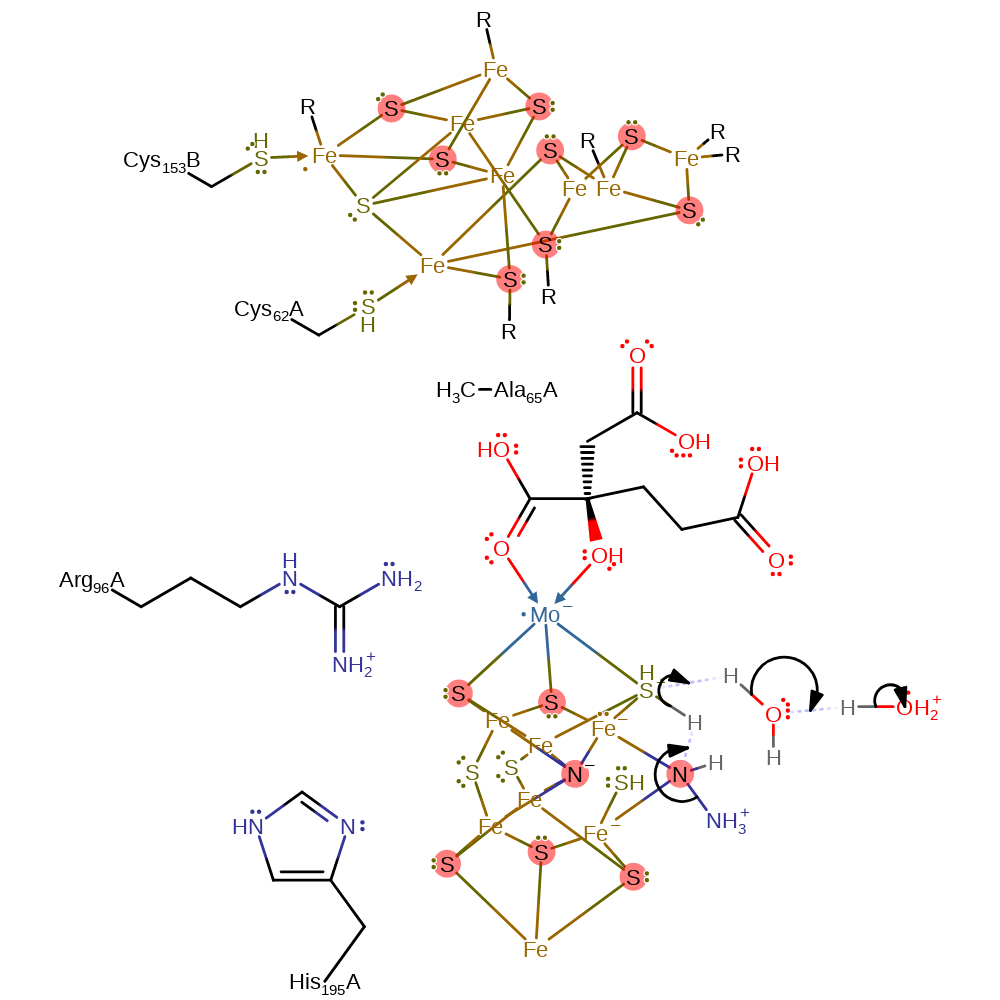
Step 11. In an elimination reaction, the N-N bond is cleaved, resulting in ammonia and a further proton being transferred from the S3B thiol, which reprotonates from water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
proton transfer, elimination (not covered by the Ingold mechanisms), intermediate formation, proton relay, overall reactant used, overall product formed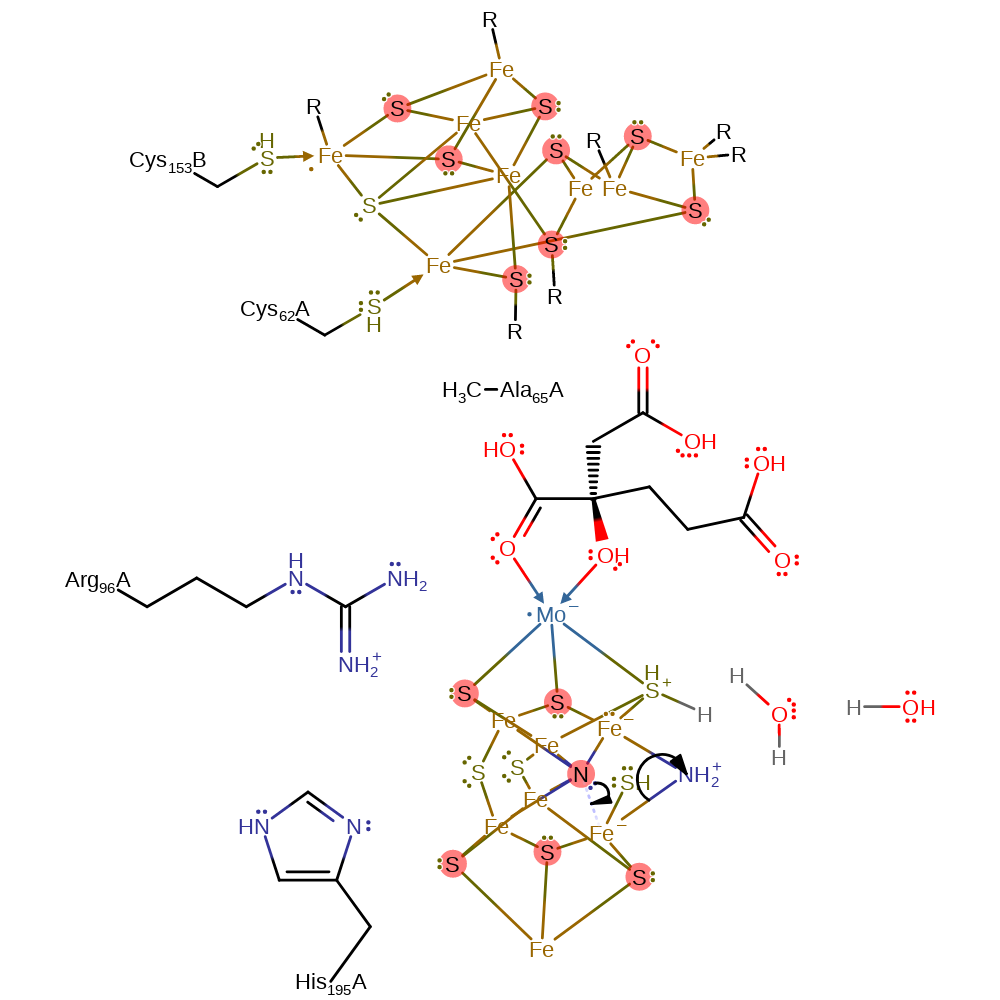
Step 12. In a nucleophilic substitution reaction the cental nitrogen atom of the cofactor reforms one of its bonds to the one of the iron centres.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
ingold: intramolecular nucleophilic substitution, intermediate formation
Step 13. The amine then deprotonates one of the thiol groups, resulting in a thiolate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
proton transfer, intermediate formation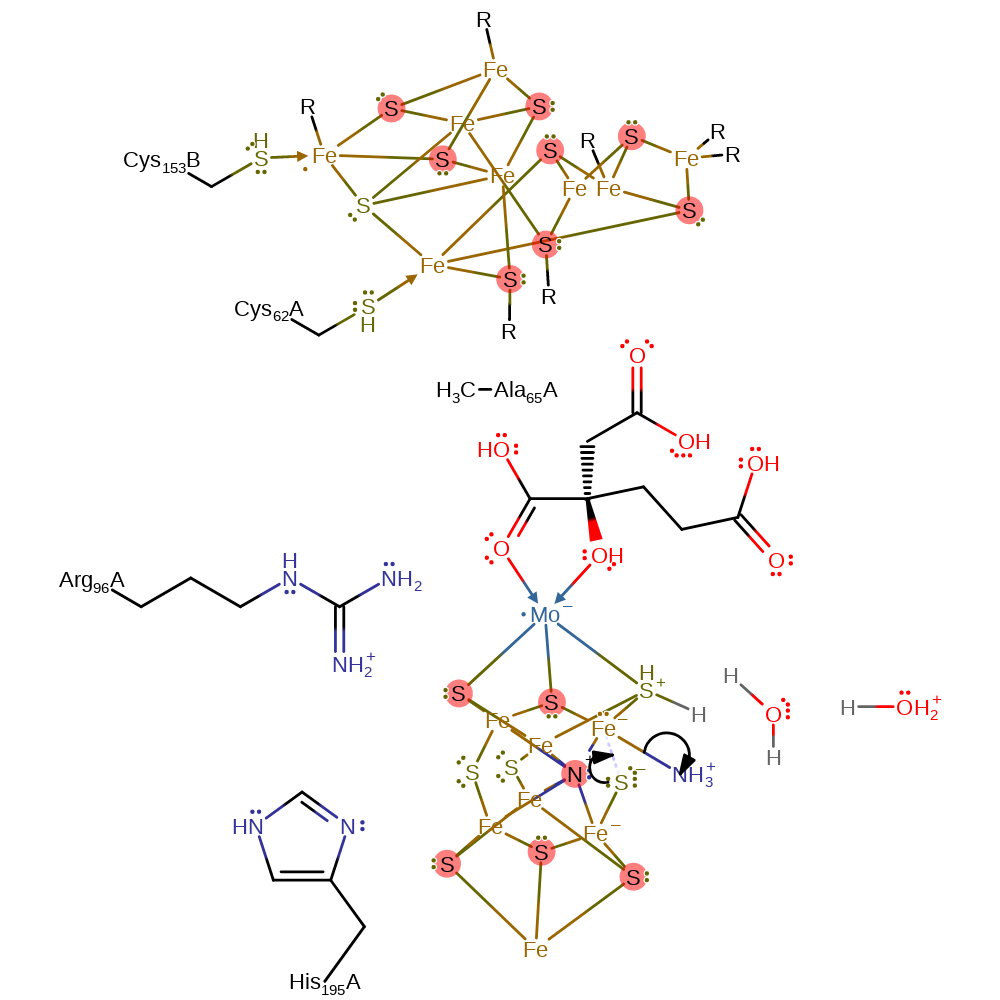
Step 14. The thiolate attacks the iron centre to which the amine group is bound in a nucleophilic substitution, producing the second ammonia.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |
Chemical Components
ingold: intramolecular nucleophilic substitution, intermediate formation, overall product formed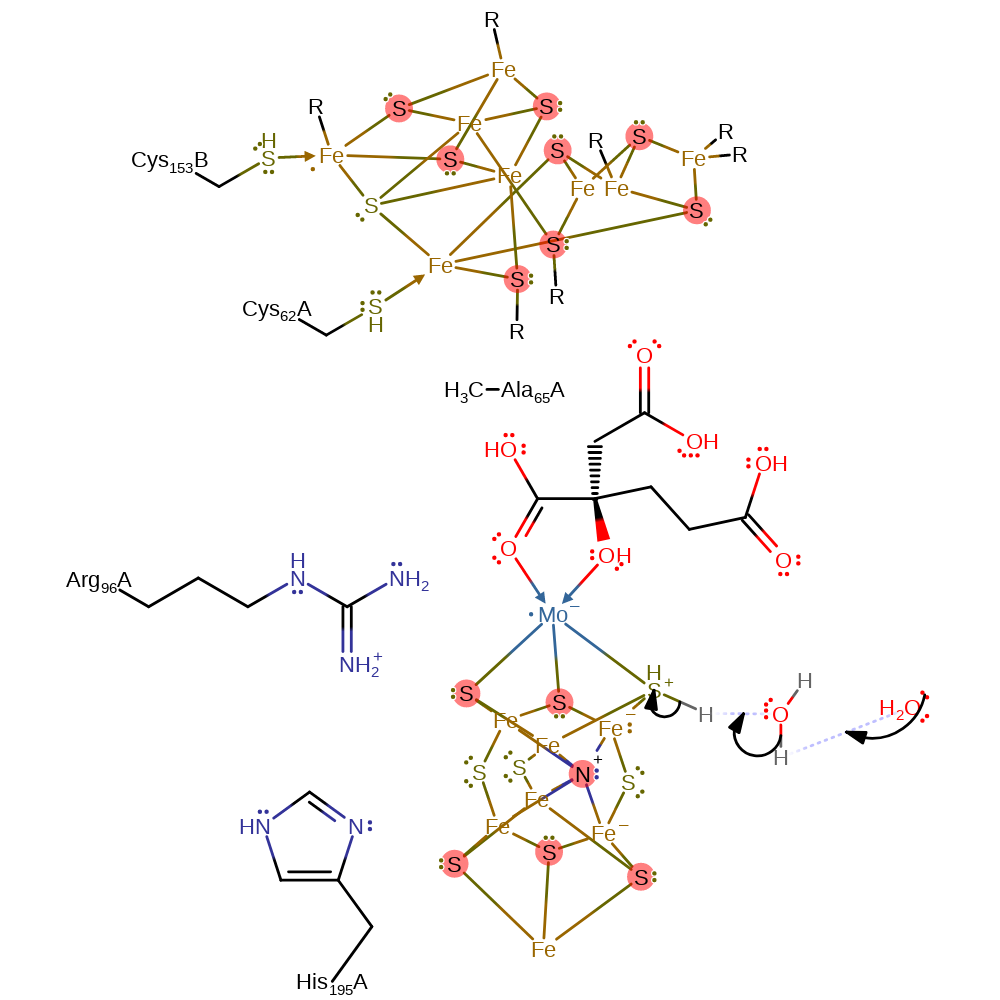
Step 15. The cofactor regenerates its initial protonation state through proton transfer.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val157(156)B (main) | polar interaction |
| Cys153(152)B | metal ligand |
| Cys62(61)A | metal ligand |
| Ala65(64)A | polar/non-polar interaction |
| Arg96(95)A | hydrogen bond donor, activator |
| His195(194)A | polar interaction, activator |










 Download:
Download: