H+-transporting two-sector ATPase (F-type, mitochondrial)
Mitochondrial membrane ATP synthase (F1F0 ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F1, containing the extramembraneous catalytic core, and F0, containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. Subunits alpha and beta form the catalytic core in F1.
F-type-ATPase utilises the proton motive force generated by photosynthesis and oxidative phosphorylation to synthesise ATP. It also catalyses the hydrolysis of ATP. During catalysis, ATP synthesis in the catalytic domain of F1 is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Rotation of the central stalk against the surrounding alpha3beta3 subunits leads to hydrolysis of ATP in three separate catalytic sites on the beta subunits.
Reference Protein and Structure
- Sequences
-
P19483

P00829 (7.1.2.2)
(7.1.2.2)
P05631
P01096
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bos taurus (Cattle)

- PDB
-
1bmf
- BOVINE MITOCHONDRIAL F1-ATPASE
(2.85 Å)



- Catalytic CATH Domains
-
3.40.50.300
 (see all for 1bmf)
(see all for 1bmf)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:7.1.2.2)
Enzyme Mechanism
Introduction
The most widely accepted mechanism of ATP synthase is the binding change mechanism. The mechanism suggests that the three catalytic sites of ATP synthase have different nucleotide binding affinities, of which one has very low substrate binding affinity, one can bind substrates reversibly and one has a very high affinity such that ATP can form spontaneously from ADP and Pi. The rotation of the central stalk, driven by the proton motive force, changes the conformation of the beta-subunits and thus the binding affinity of the 3 catalytic sites, taking each through cycles of the 3 affinity states and hence catalysing the synthesis and hydrolysis of ATP. Though, it is worthwhile to note that there are still disagreements on some of the proposals in the mechanism. The transition state of the phosphoryl transfer is general accepted as a pentacovalent phosphorus with 2 apical and 3 equatorial bonds. This transition state of the phosphoryl transfer is stabilised by 3 residues, alpha-Arg373, beta-Lys162 and beta-Arg189. In hydrolysis of ATP, beta-Glu188 polarises and activates a deprotonated water molecule for an inline nucleophilic attack on the terminal-phosphate of the ATP.
Catalytic Residues Roles
| UniProt | PDB* (1bmf) | ||
| Glu238 | Glu188(192)D | Acts as a base to deprotonate, thus activating a water molecule to allow its nucleophilic attack on the terminal phosphate of ATP in hydrolysis of ATP. | electrostatic stabiliser |
| Lys212, Arg239, Arg416 | Lys162(166)D, Arg189(193)D, Arg373C | Stabilises the pentacovalent negatively charged phosphorus transition state. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, intermediate formation, proton relay, rate-determining step, unimolecular elimination by the conjugate base, intermediate collapse, intermediate terminated, overall product formed, dephosphorylationReferences
- Dittrich M et al. (2004), Biophys J, 87, 2954-2967. ATP Hydrolysis in the βTP and βDP Catalytic Sites of F1-ATPase. DOI:10.1529/biophysj.104.046128. PMID:15315950.
- Watanabe R et al. (2014), J Biol Chem, 289, 19331-19340. Robustness of the rotary catalysis mechanism of F1-ATPase. DOI:10.1074/jbc.M114.569905. PMID:24876384.
- Martín-García F et al. (2013), Biochemistry, 52, 959-966. Simulation of catalytic water activation in mitochondrial F1-ATPase using a hybrid quantum mechanics/molecular mechanics approach: an alternative role for β-Glu 188. DOI:10.1021/bi301109x. PMID:23320924.
- Senior AE et al. (2002), Biochim Biophys Acta, 1553, 188-211. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. DOI:10.1016/s0005-2728(02)00185-8. PMID:11997128.
- Menz RI et al. (2001), Cell, 106, 331-341. Faculty of 1000 evaluation for Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. DOI:10.3410/f.1001775.15458. PMID:11509182.
- Gibbons C et al. (2000), Nat Struct Biol, 7, 1055-1061. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. DOI:10.1038/80981. PMID:11062563.
- Nadanaciva S et al. (1999), Biochemistry, 38, 15493-15499. Importance of F1-ATPase Residue α-Arg-376 for Catalytic Transition State Stabilization†. DOI:10.1021/bi9917683. PMID:10569931.
- Nadanaciva S et al. (1999), Biochemistry, 38, 7670-7677. The Role of β-Arg-182, an Essential Catalytic Site Residue inEscherichia coliF1-ATPase. DOI:10.1021/bi990663x. PMID:10387006.
- Löbau S et al. (1997), J Biol Chem, 272, 3648-3656. F1-ATPase, Roles of Three Catalytic Site Residues. DOI:10.1074/jbc.272.6.3648. PMID:9013618.
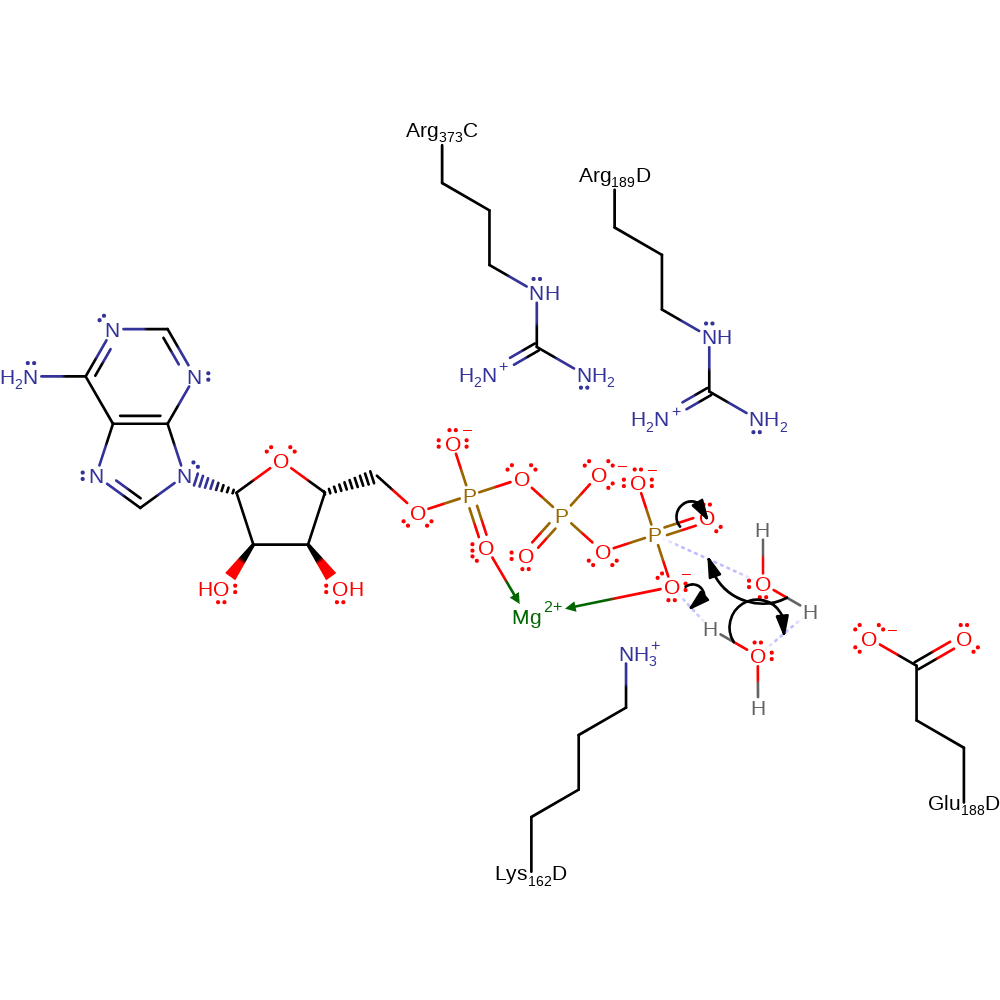
Step 1. The gamma phopshate of ATP deprotonates the first water, which deprotonates a second water, which attacks the gamma phosphate in a nucleophilic addition resulting in a pentavalent phosphate intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg373C | electrostatic stabiliser |
| Lys162(166)D | electrostatic stabiliser |
| Glu188(192)D | electrostatic stabiliser |
| Arg189(193)D | electrostatic stabiliser |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation, proton relay, rate-determining step
Step 2. The pentavalent phosphate collapses, releasing orthophosphate and ADP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg373C | electrostatic stabiliser |
| Lys162(166)D | electrostatic stabiliser |
| Glu188(192)D | electrostatic stabiliser |
| Arg189(193)D | electrostatic stabiliser |





 Download:
Download: