Purine-nucleoside phosphorylase
Purine-nucleoside phosphorylase (PNP) catalyses the reversible phosphorolysis of purine nucleosides to generate the corresponding purine base and ribose 1-phosphate. There are two types of this enzyme, showing reasonably high sequence similarity within families and little identity between them. Type I (of which this entry is an example) tend to be trimeric and are found mainly in mammals. Type I enzymes are specific for inosine/guanine nucleosides and have a molecular mass of around 90kDa. Type II tend to be hexameric (although some are thought to be tetramers) and have broad substrate specificity. Type II enzymes are found mainly in prokaryotes and have a molecular mass between 110 and 150 kDa. There are also some PNP enzymes which fall into neither class.
Reference Protein and Structure
- Sequence
-
P00491
 (2.4.2.1)
(2.4.2.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1rr6
- Structure of human purine nucleoside phosphorylase in complex with Immucillin-H and phosphate
(2.5 Å)



- Catalytic CATH Domains
-
3.40.50.1580
 (see all for 1rr6)
(see all for 1rr6)
Enzyme Reaction (EC:2.4.2.1)
Enzyme Mechanism
Introduction
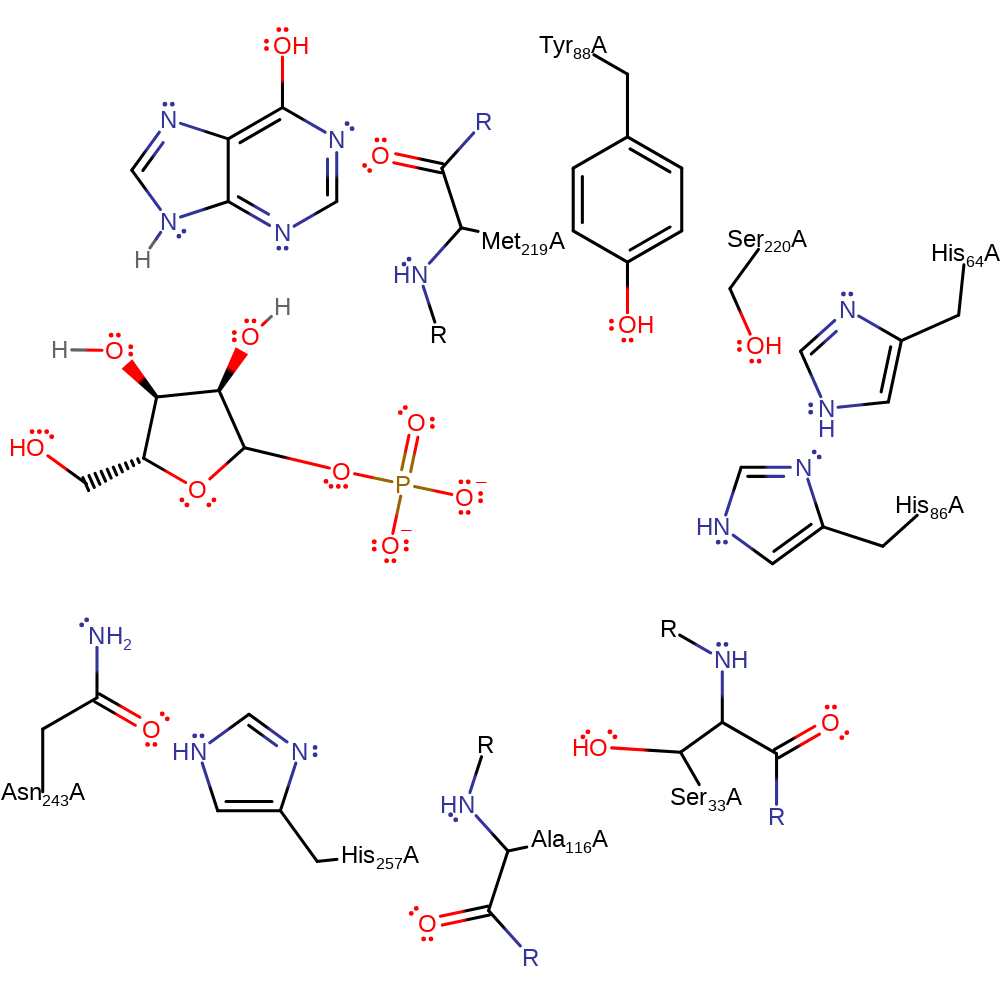
The phosphate group is di-anionic and is stabilised by the Ser33-His64-His86 catalytic triad. It attacks the electrophilic C1 involved in the glycosidic bond in an SN2 manner. His257, Tyr88, Met119 all stabilise the ribose ring transition state while Ala116 and Ser220 also stabilise alongside the triad the phosphate transition state. Hypoxanthine is then protonated by the phosphoryl proton via the 2' and 3' OH groups on the ribose, although there are multiple pathways that protonation can take place through.
Catalytic Residues Roles
| UniProt | PDB* (1rr6) | ||
| Ser220 | Ser220A | Stabilise phosphate by a side chain hydrogen bond. | hydrogen bond donor, electrostatic stabiliser |
| Ala116 (main-N), Ser33 (main-N) | Ala116A (main-N), Ser33A (main-N) | Stabilise and position the phosphate group through their main chain amide groups. | hydrogen bond donor, electrostatic stabiliser |
| Tyr88, His257, Met219 (main-N) | Tyr88A, His257A, Met219A (main-N) | Stabilise the ribose ring in the transition state via hydrogen bonds. | hydrogen bond donor, electrostatic stabiliser |
| His64, Ser33, His86 | His64A, Ser33A, His86A | Catalytic triad stabilises the nucleophilic phosphate. | electrostatic stabiliser |
| Asn243 | Asn243A | Stabilises base via hydrogen bonding, thought to also be involved in determining substrate specificity. | electrostatic stabiliser, polar interaction |
Chemical Components
bimolecular nucleophilic substitution, rate-determining step, heterolysis, overall reactant used, proton relay, overall product formed, proton transferReferences
- Isaksen GV et al. (2016), Biochemistry, 55, 2153-2162. Computer Simulations Reveal Substrate Specificity of Glycosidic Bond Cleavage in Native and Mutant Human Purine Nucleoside Phosphorylase. DOI:10.1021/acs.biochem.5b01347. PMID:26985580.
- Shi W et al. (2004), J Biol Chem, 279, 18103-18106. Plasmodium falciparum purine nucleoside phosphorylase: crystal structures, immucillin inhibitors, and dual catalytic function. DOI:10.1074/jbc.C400068200. PMID:14982926.
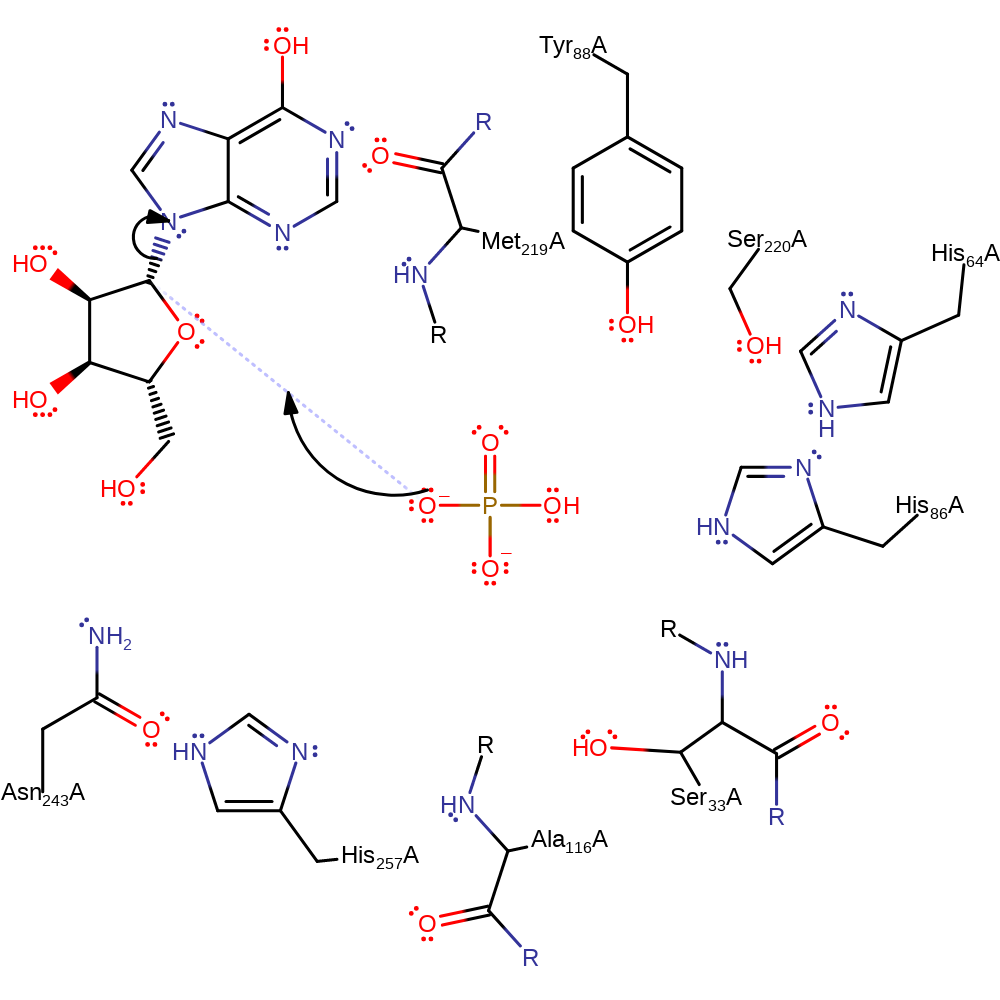
Step 1. The incoming nucleophilic phosphate group is stabilised by the Ser33-His64-His86 catalytic triad. O4 on the phosphate attacks the glycosidic C1.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His86A | hydrogen bond acceptor |
| Ser33A | electrostatic stabiliser |
| His64A | electrostatic stabiliser |
| Asn243A | electrostatic stabiliser |
| Met219A (main-N) | electrostatic stabiliser |
| Ser220A | electrostatic stabiliser, hydrogen bond donor |
| Tyr88A | electrostatic stabiliser |
| His257A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr88A | hydrogen bond donor |
| Met219A (main-N) | hydrogen bond donor |
| Ala116A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser33A | hydrogen bond donor |
| Ser33A (main-N) | hydrogen bond donor |
| Asn243A | polar interaction |
Chemical Components
ingold: bimolecular nucleophilic substitution, rate-determining step, heterolysis, overall reactant used
Step 2. Proton shuttle from the phosphate group via the 2' and 3' OH on the ribose sugar to N9 on hypoxanthine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
proton relay, overall product formed, proton transferIntroduction
Proposed mechanism in which the phosphate is activated by His86 and attacks the ribose ring directly via an SN2 type reaction.
Catalytic Residues Roles
| UniProt | PDB* (1rr6) | ||
| His86 | His86A | Acts as a general acid/base, abstracting a proton from the phosphate substrate and being returned to its initial protonation state by the purine product. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu89 | Glu89A | Activates the catalytic His. | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Asn243 | Asn243A | Stabilises the negatively charged intermediate. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, intermediate formation, heterolysis, charge delocalisation, intermediate collapse, overall product formed, native state of enzyme regenerated, intermediate terminatedReferences
- Canduri F et al. (2005), Biochem Biophys Res Commun, 327, 646-649. New catalytic mechanism for human purine nucleoside phosphorylase. DOI:10.1016/j.bbrc.2004.12.052. PMID:15649395.
- Craig SP 3rd et al. (2000), J Biol Chem, 275, 20231-20234. Purine Phosphoribosyltransferases. DOI:10.1074/jbc.r000002200. PMID:10816600.
- Mao C et al. (1998), Biochemistry, 37, 7135-7146. Calf Spleen Purine Nucleoside Phosphorylase Complexed with Substrates and Substrate Analogues†,‡. DOI:10.1021/bi9723919. PMID:9585525.
- Koellner G et al. (1997), J Mol Biol, 265, 202-216. Crystal structure of calf spleen purine nucleoside phosphorylase in a complex with hypoxanthine at 2.15 Å resolution. DOI:10.1006/jmbi.1996.0730. PMID:9020983.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu89A | hydrogen bond acceptor, activator |
| His86A | hydrogen bond donor, hydrogen bond acceptor |
| Asn243A | hydrogen bond donor |
| His86A | proton acceptor |
Chemical Components
proton transfer, overall reactant used, intermediate formation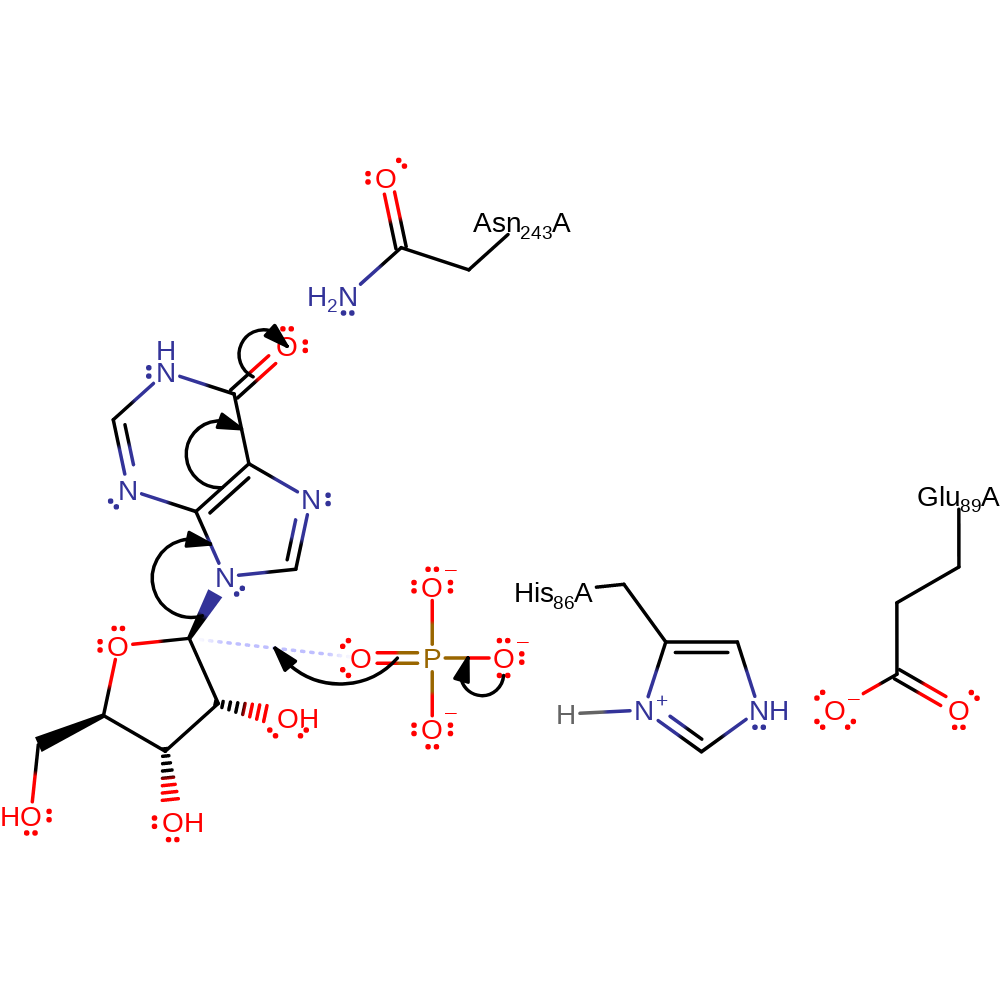
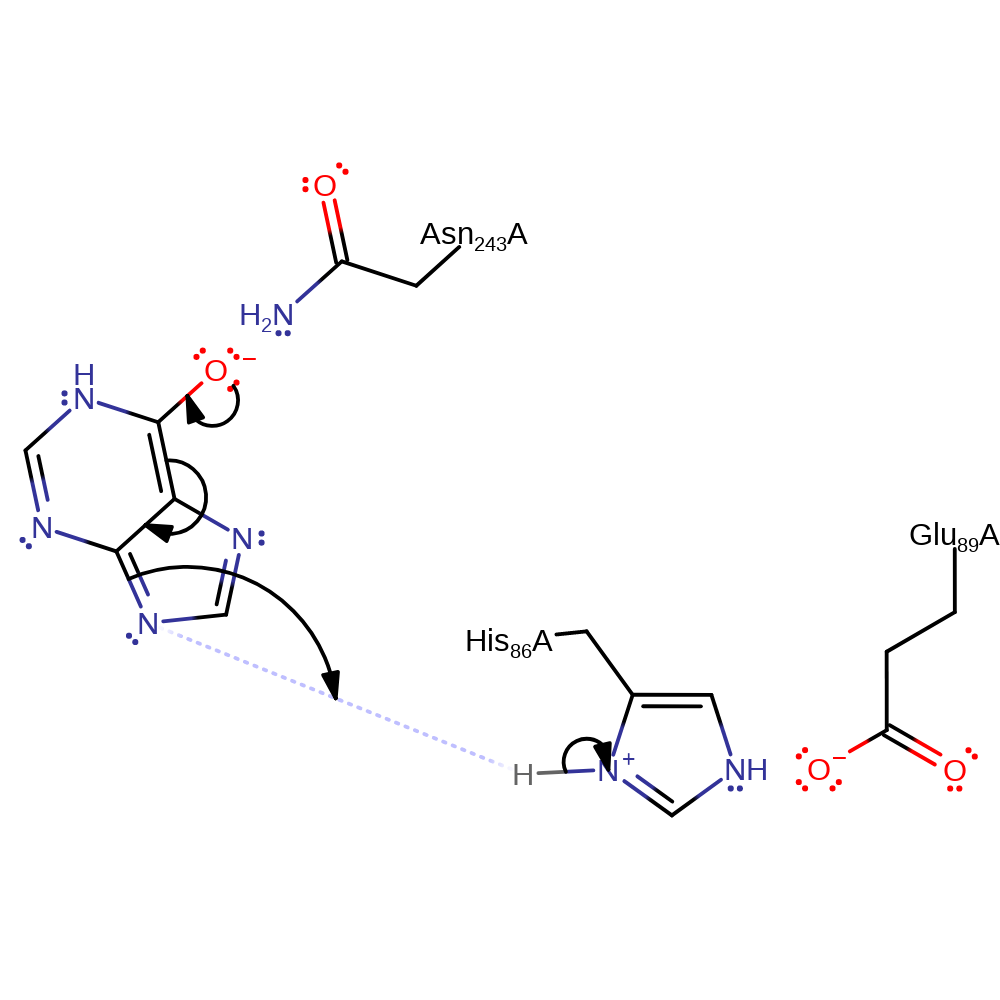
Step 2. The phosphate initiates a nucleophilic attack on the substrate, resulting in C-N bond cleavage.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu89A | hydrogen bond acceptor, electrostatic stabiliser |
| His86A | hydrogen bond donor |
| Asn243A | electrostatic stabiliser |
Chemical Components
heterolysis, overall reactant used, charge delocalisation, intermediate collapseCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu89A | hydrogen bond acceptor |
| His86A | hydrogen bond donor |
| Asn243A | electrostatic stabiliser, hydrogen bond donor |
| His86A | proton donor |
Chemical Components
proton transfer, overall product formed, native state of enzyme regenerated, intermediate terminatedIntroduction
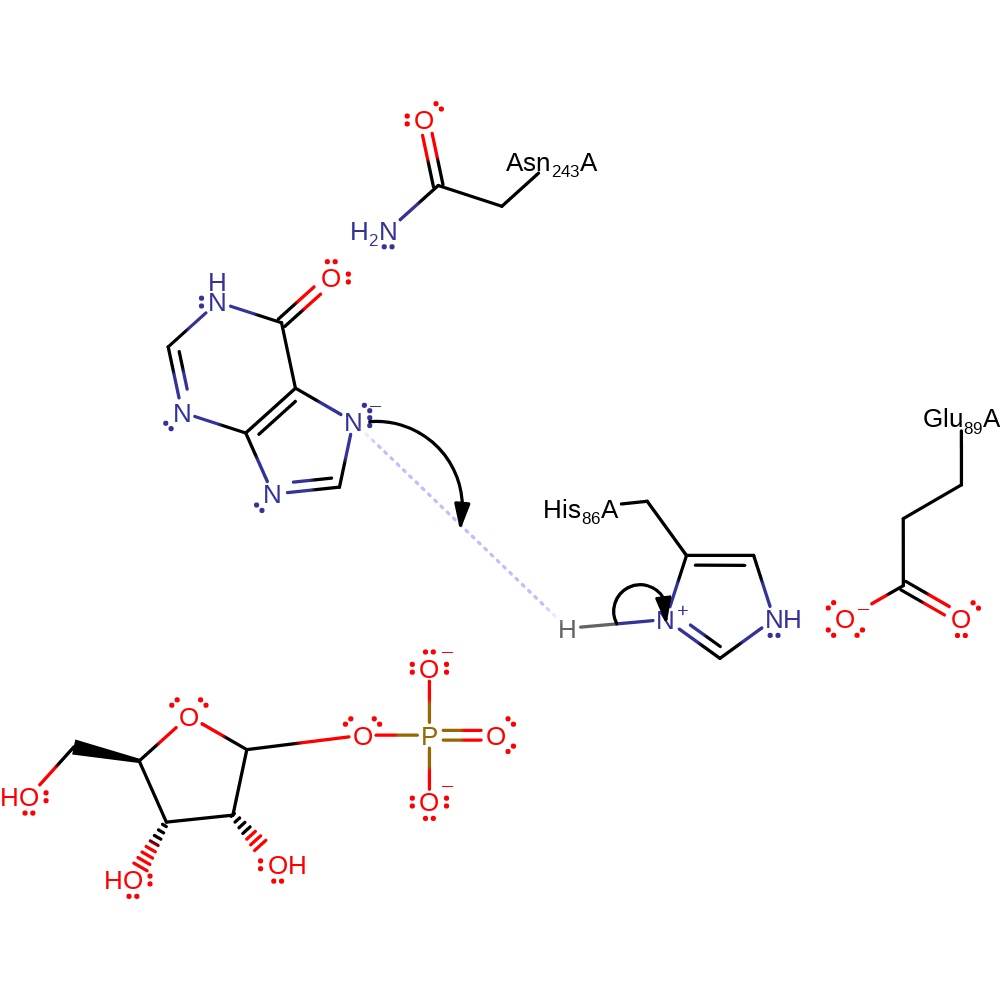
The substitution reaction occurs by a SN1 type mechanism. His86 is hydrogen bonded to Glu89, and abstracts a proton from the phosphate ion which causes strain and weakening of the glycosidic bond. The glycosidic bond is cleaved, forming an oxycarbenium ion intermediate which is stabilised by the phosphate dianion. Therefore, the phosphate anion is used for both the initiation of bond cleavage and stabilisation of intermediate [PMID:9305963].
Catalytic Residues Roles
| UniProt | PDB* (1rr6) | ||
| His86 | His86A | Acts as a general acid/base, abstracting a proton from the substrate phosphate. It is returned to its initial protonation state by the purine product. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Glu89 | Glu89A | Activates the catalytic His. | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Asn243 | Asn243A | Helps stabilise the negatively charged intermediates. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, intermediate formation, heterolysis, charge delocalisation, elimination (not covered by the Ingold mechanisms), bimolecular nucleophilic addition, overall product formed, intermediate terminated, native state of enzyme regeneratedReferences
- Erion MD et al. (1997), Biochemistry, 36, 11735-11748. Purine Nucleoside Phosphorylase. 2. Catalytic Mechanism†. DOI:10.1021/bi961970v. PMID:9305963.
- Isaksen GV et al. (2016), Biochemistry, 55, 2153-2162. Computer Simulations Reveal Substrate Specificity of Glycosidic Bond Cleavage in Native and Mutant Human Purine Nucleoside Phosphorylase. DOI:10.1021/acs.biochem.5b01347. PMID:26985580.
- Canduri F et al. (2005), Biochem Biophys Res Commun, 327, 646-649. New catalytic mechanism for human purine nucleoside phosphorylase. DOI:10.1016/j.bbrc.2004.12.052. PMID:15649395.
- Mao C et al. (1998), Biochemistry, 37, 7135-7146. Calf Spleen Purine Nucleoside Phosphorylase Complexed with Substrates and Substrate Analogues†,‡. DOI:10.1021/bi9723919. PMID:9585525.
- Koellner G et al. (1998), J Mol Biol, 280, 153-166. Crystal structure of the ternary complex of E. coli purine nucleoside phosphorylase with formycin B, a structural analogue of the substrate inosine, and phosphate (sulphate) at 2.1 Å resolution. DOI:10.1006/jmbi.1998.1799. PMID:9653038.
- Koellner G et al. (1997), J Mol Biol, 265, 202-216. Crystal structure of calf spleen purine nucleoside phosphorylase in a complex with hypoxanthine at 2.15 Å resolution. DOI:10.1006/jmbi.1996.0730. PMID:9020983.
- Erion MD et al. (1997), Biochemistry, 36, 11725-11734. Purine Nucleoside Phosphorylase. 1. Structure−Function Studies†. DOI:10.1021/bi961969w. PMID:9305962.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu89A | hydrogen bond acceptor, activator |
| His86A | hydrogen bond donor, hydrogen bond acceptor |
| Asn243A | hydrogen bond donor |
| His86A | proton acceptor |
Chemical Components
proton transfer, overall reactant used, intermediate formation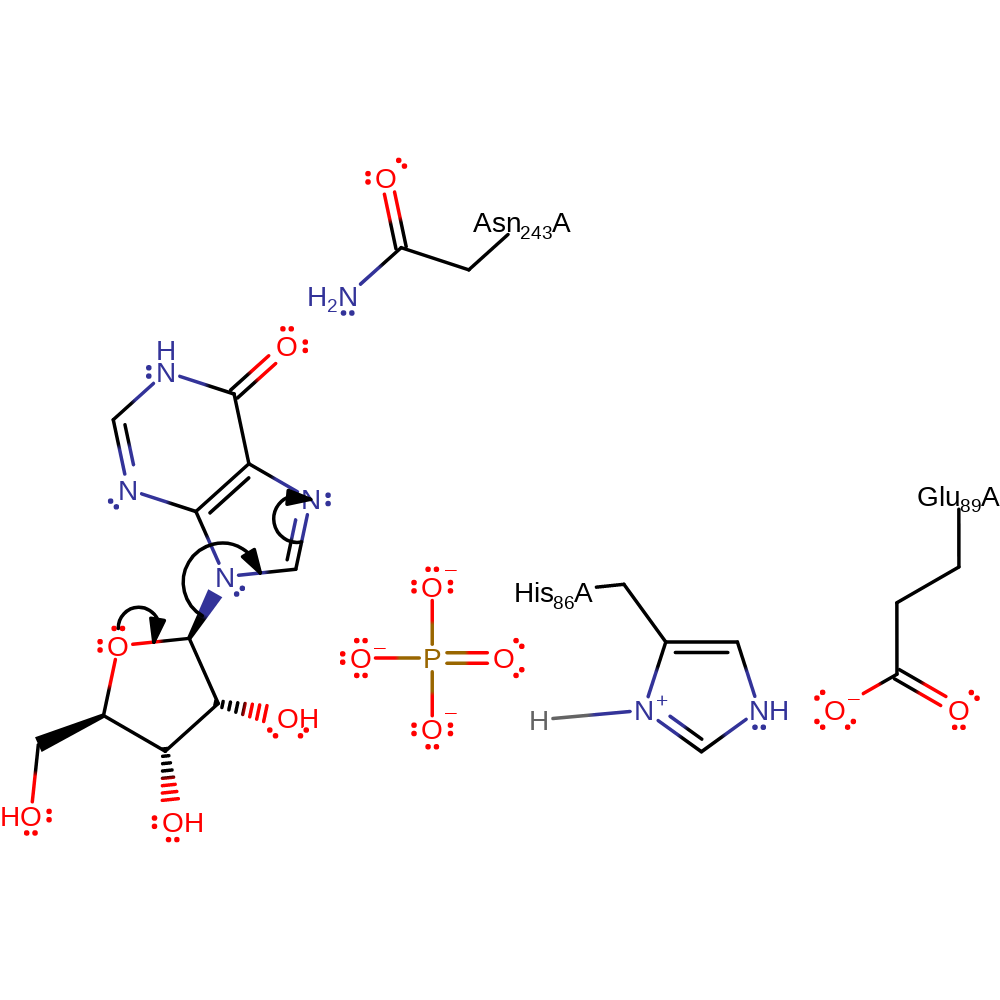
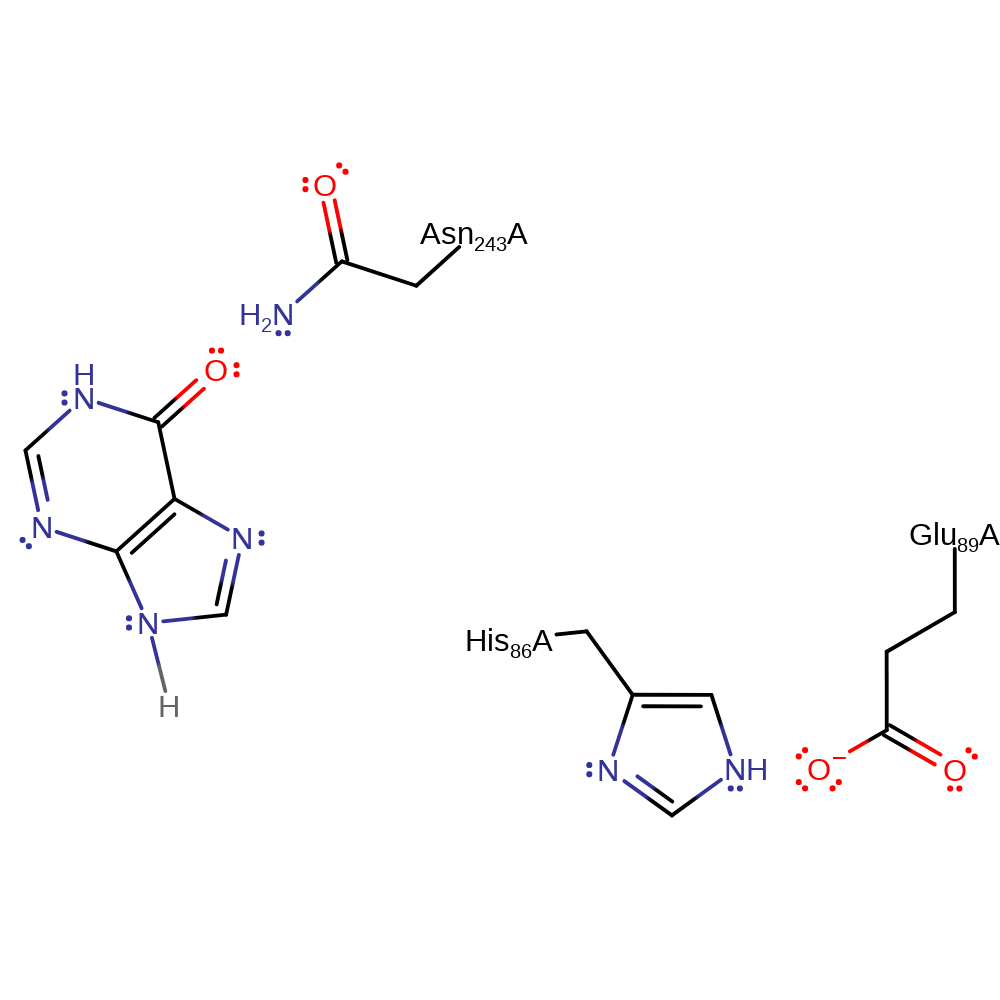
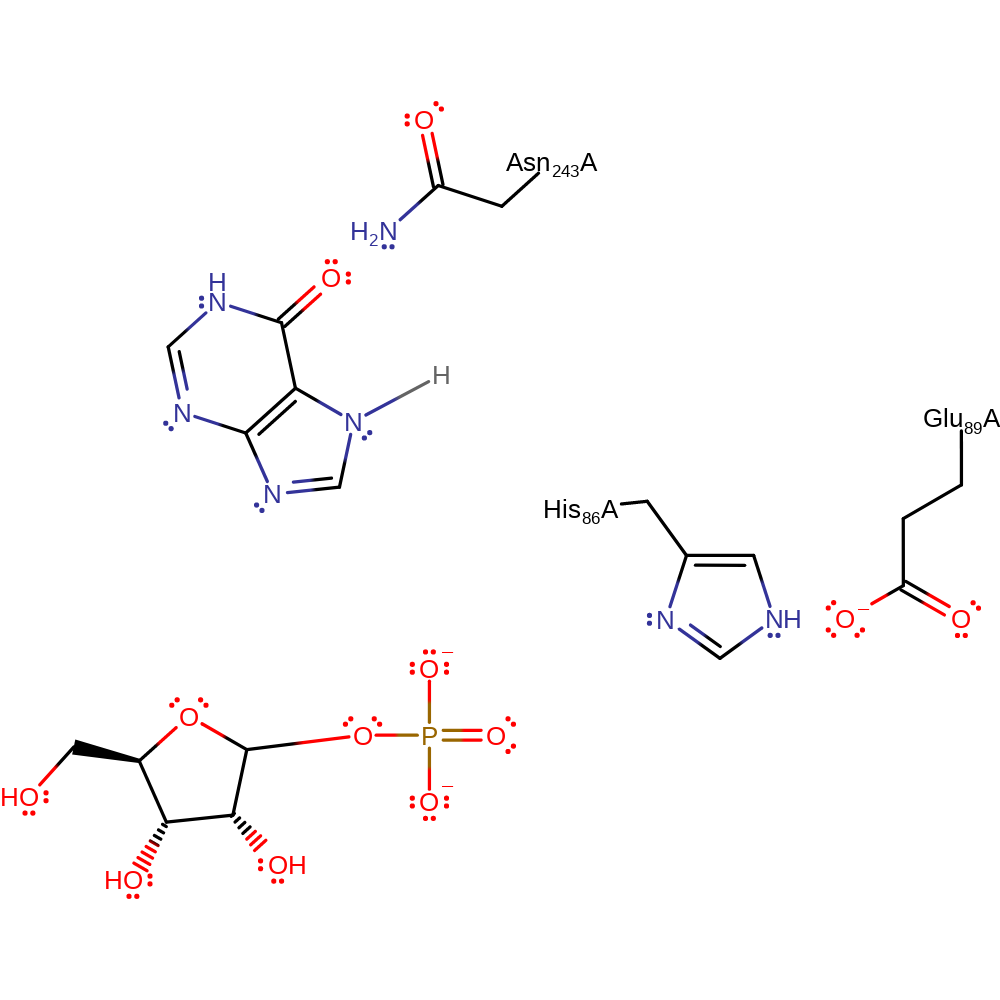
Step 2. In a heterolysis reaction, the C-N bond between ribose and the purine is cleaved.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu89A | hydrogen bond acceptor, electrostatic stabiliser |
| His86A | hydrogen bond donor |
| Asn243A | electrostatic stabiliser |
Chemical Components
heterolysis, overall reactant used, charge delocalisation, intermediate formation, elimination (not covered by the Ingold mechanisms)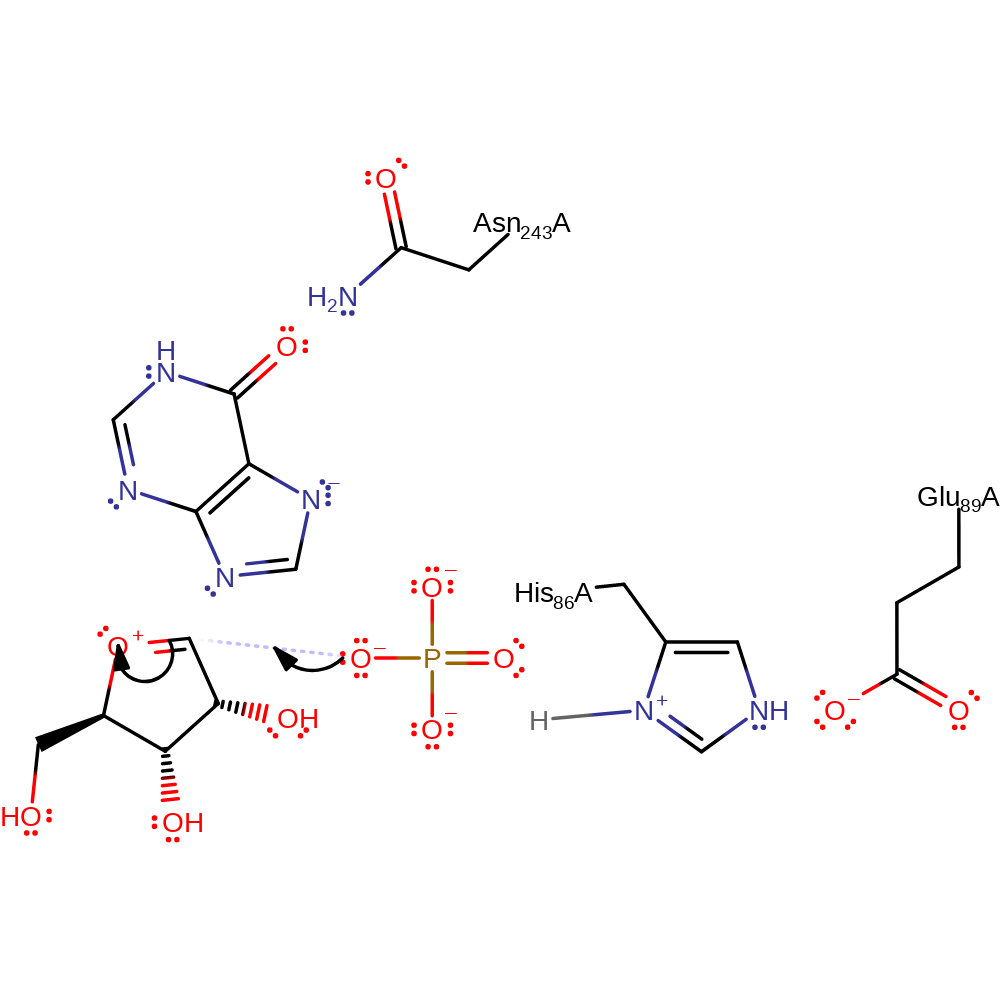
Step 3. Phosphate initiates a nucleophilic attack on the ribose in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu89A | hydrogen bond acceptor, electrostatic stabiliser |
| His86A | hydrogen bond donor, electrostatic stabiliser |
| Asn243A | electrostatic stabiliser, hydrogen bond donor |
Chemical Components
ingold: bimolecular nucleophilic addition, overall product formed, intermediate terminatedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu89A | hydrogen bond acceptor |
| His86A | hydrogen bond donor |
| Asn243A | electrostatic stabiliser, hydrogen bond donor |
| His86A | proton donor |




 Download:
Download: 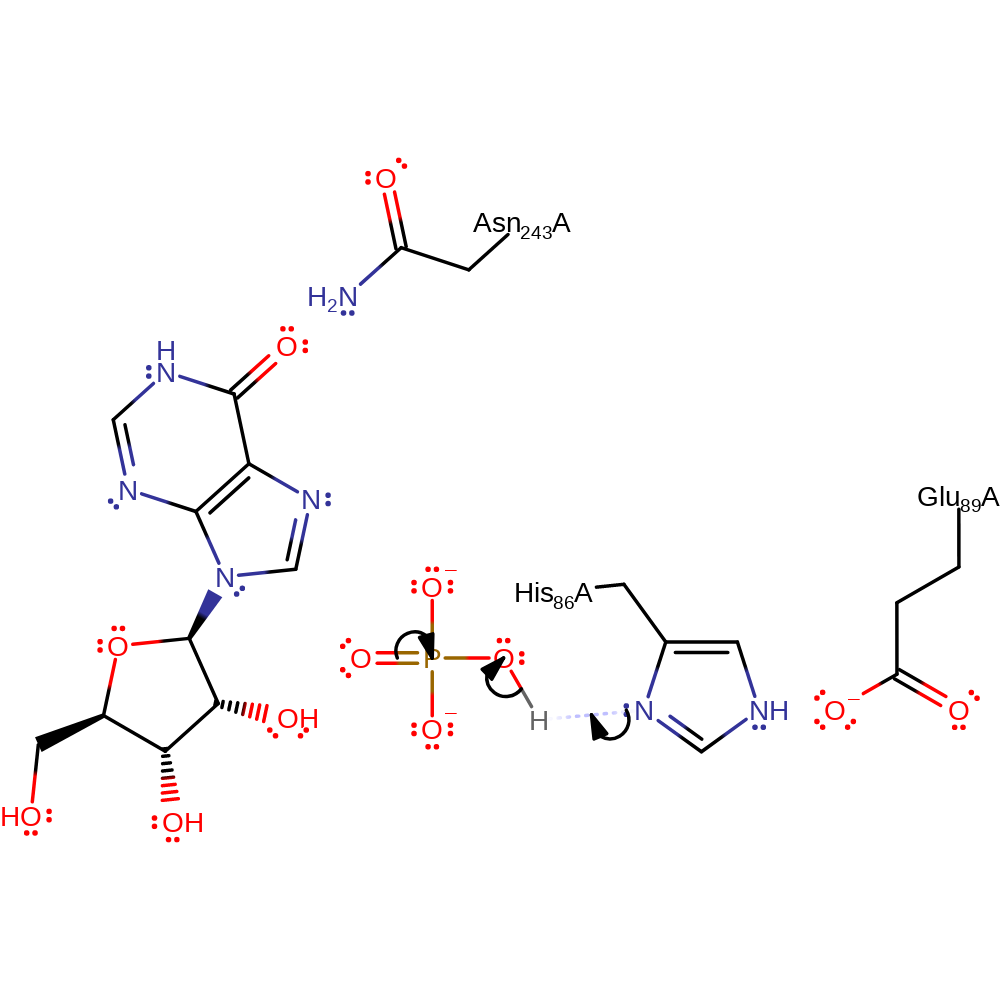
 Download:
Download: 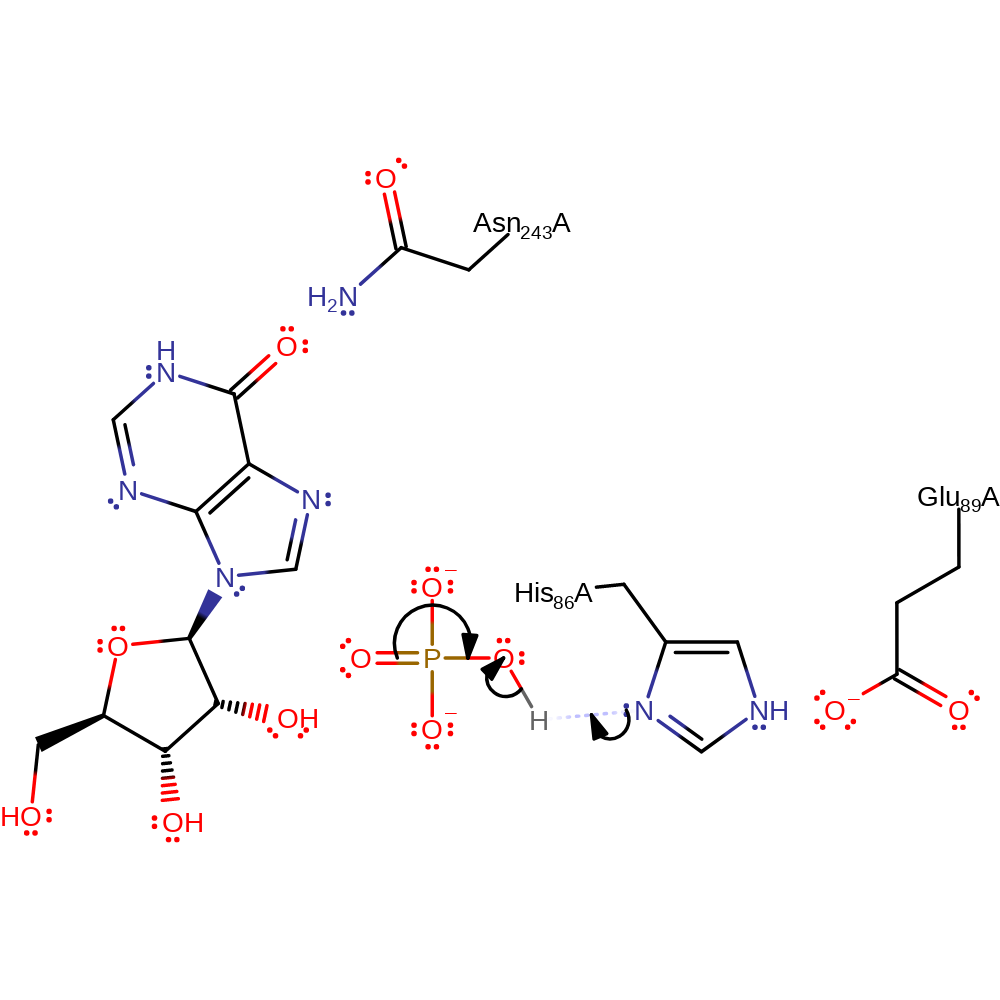
 Download:
Download: