Chloride peroxidase (vanadate dependent)
Chloride peroxidase oxidises chloride in the presence of hydrogen peroxidase to form hypochlorous acid. This acid is the final product of the reaction in the absence of a suitable nucleophilic acceptor. However, it can go on to form a diversity of halogenated reaction products if a convenient nucleophilic acceptor is present.
Hypochlorous acid is strongly bacteriocidal and a powerful oxidising agent. It is secreted by pathogenic fungi e.g. Curvularia inaequalis in a mechanism to oxidise plant cell walls to facilitate penetration of the fungus into the host amongst other uses. It can also act upon Br- and I-.
Reference Protein and Structure
- Sequence
-
P49053
 (1.11.1.10)
(1.11.1.10)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Curvularia inaequalis (Fungus)

- PDB
-
1vnc
- CHLOROPEROXIDASE FROM THE FUNGUS CURVULARIA INAEQUALIS
(2.1 Å)



- Catalytic CATH Domains
-
1.10.606.10
 (see all for 1vnc)
(see all for 1vnc)
- Cofactors
- Vanadate(3-) (1) Metal MACiE
Enzyme Reaction (EC:1.11.1.10)
Enzyme Mechanism
Introduction
The enzyme contains vanadium in the active site. Vanadium has a trigonal bipyramid structure and is coordinated by three non protein oxygen ligands, His496 and an exogenous azide ligand. The negative charge is compensated for by hydrogen bonds to Arg360, Arg490, Lys353, Gly403 and Ser402. Catalysis is thought to occur through a ping-pong mechanism. The first step is the binding of the hydrogen peroxide at the same binding site, His404, as the azide to form a peroxo intermediate. The Cl- then binds the activated peroxidase. This is stabilised by hydrophobic interactions with Trp350, Phe397 and the imidazole ring of His404. Without knowledge of the intermediates formed the exact mechanism remains unknown. His404 is thought to play a crucial role as an acid-base group as changes in pH inhibit the reaction. Nucleophilic attack of Cl- on the activated peroxide or halide oxidation through the coordinated metal would allow the reaction to proceed.
Catalytic Residues Roles
| UniProt | PDB* (1vnc) | ||
| His404 | His404A | Helps stabilise the activated peroxidase and likely acts as a general acid/base throughout the reaction. | activator, hydrogen bond acceptor |
| His496 | His496A | Binds the vanadate cofactor. | covalently attached, metal ligand |
| Arg360, Ser402, Arg490, Gly403 (main-N), Lys353 | Arg360A, Ser402A, Arg490A, Gly403A (main-N), Lys353A | Form hydrogen bonds to the vanadate cofactor, compensating for the high concentration of negative charge present. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, intermediate formation, cofactor used, acidic bimolecular nucleophilic substitution, intermediate collapse, overall product formed, dehydration, coordination to a metal ion, decoordination from a metal ion, intramolecular nucleophilic substitution, cyclisation, coordination, intramolecular elimination, substitution (not covered by the Ingold mechanisms), intermediate terminated, decyclisation, native state of cofactor regenerated, rate-determining stepReferences
- Messerschmidt A et al. (1997), Biol Chem, 378, 309-315. Implications for the Catalytic Mechanism of the Vanadium-Containing Enzyme Chloroperoxidase from the Fungus Curvularia inaequalis by X-Ray Structures of the Native and Peroxide Form. DOI:10.1515/bchm.1997.378.3-4.309. PMID:9165086.
- Frank A et al. (2016), Chembiochem, 17, 2028-2032. Characterization of a Cyanobacterial Haloperoxidase and Evaluation of its Biocatalytic Halogenation Potential. DOI:10.1002/cbic.201600417. PMID:27542168.
- Gupta R et al. (2015), J Am Chem Soc, 137, 5618-5628. 51V NMR Crystallography of Vanadium Chloroperoxidase and Its Directed Evolution P395D/L241V/T343A Mutant: Protonation Environments of the Active Site. DOI:10.1021/jacs.5b02635. PMID:25856001.
- Fournier JB et al. (2014), Appl Environ Microbiol, 80, 7561-7573. The Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Species Zobellia galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in the Vanadium Haloperoxidase Enzyme Family. DOI:10.1128/aem.02430-14. PMID:25261522.
- Winter JM et al. (2009), J Biol Chem, 284, 18577-18581. Exploring the Chemistry and Biology of Vanadium-dependent Haloperoxidases. DOI:10.1074/jbc.r109.001602. PMID:19363038.
- Schneider CJ et al. (2008), J Am Chem Soc, 130, 2712-2713. Elucidating the Protonation Site of Vanadium Peroxide Complexes and the Implications for Biomimetic Catalysis. DOI:10.1021/ja077404c. PMID:18266364.
- Zampella G et al. (2006), Inorg Chem, 45, 7133-7143. Insight into the Catalytic Mechanism of Vanadium Haloperoxidases. DFT Investigation of Vanadium Cofactor Reactivity. DOI:10.1021/ic060555g. PMID:16933914.
- Bortolini O et al. (2005), J Inorg Biochem, 99, 1549-1557. Vanadium (V) peroxocomplexes: Structure, chemistry and biological implications. DOI:10.1016/j.jinorgbio.2005.04.003. PMID:15964077.
- Hemrika W et al. (1999), J Biol Chem, 274, 23820-23827. Heterologous Expression of the Vanadium-containing Chloroperoxidase from Curvularia inaequalis inSaccharomyces cerevisiae and Site-directed Mutagenesis of the Active Site Residues His496, Lys353, Arg360, and Arg490. DOI:10.1074/jbc.274.34.23820. PMID:10446144.
- Messerschmidt A et al. (1996), Proc Natl Acad Sci U S A, 59, 580-396. X-ray structure of a vanadium-containing chloroperoxidase from the fungus Curvularia inaequalis. DOI:10.1016/0162-0134(95)97673-e. PMID:8552646.

Step 1. The axial hydroxide of the vanadate cofactor deprotonates hydrogen peroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His404A | hydrogen bond acceptor, activator |
| Lys353A | hydrogen bond donor, electrostatic stabiliser |
| Arg360A | hydrogen bond donor, electrostatic stabiliser |
| Arg490A | hydrogen bond donor, electrostatic stabiliser |
| Ser402A | hydrogen bond donor, electrostatic stabiliser |
| Gly403A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| His496A | covalently attached, metal ligand |
Chemical Components
proton transfer, overall reactant used, intermediate formation, cofactor used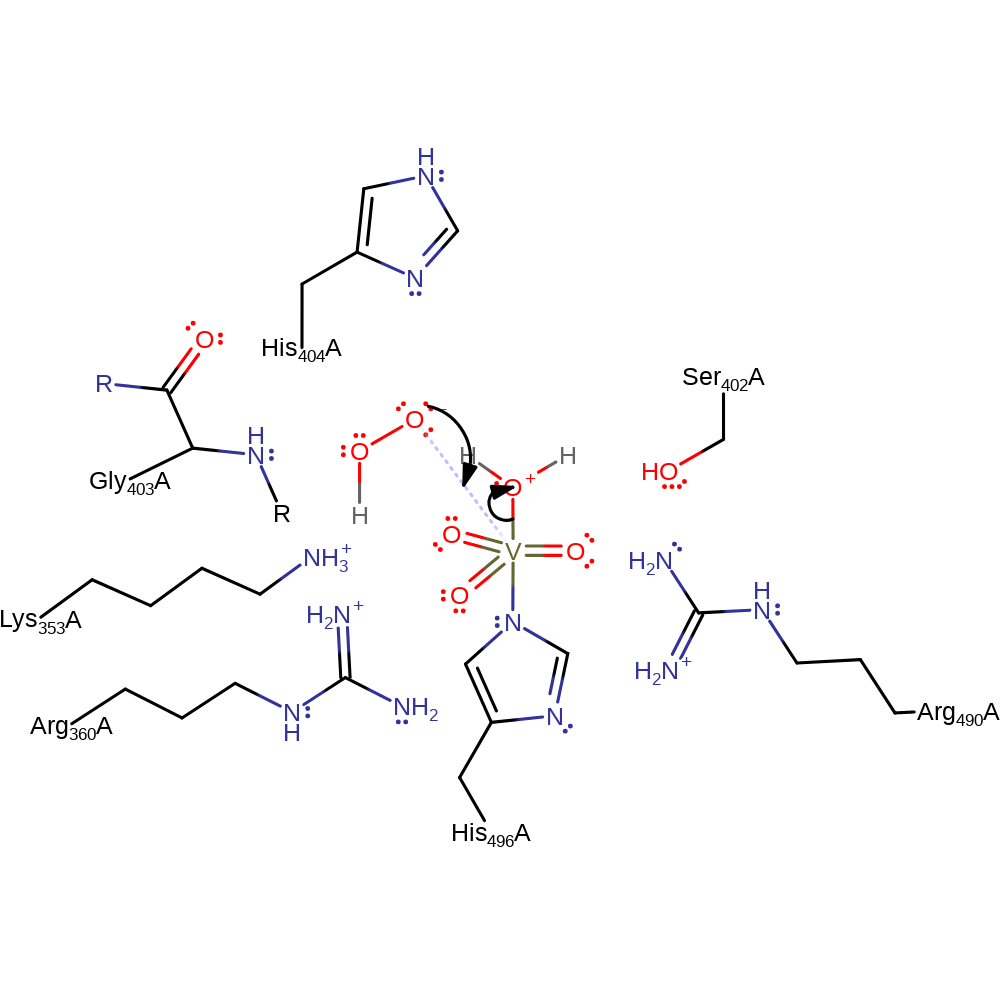
Step 2. The activated hydrogen peroxide initiates a nucleophilic attack on the vanadate in a substitution reaction, eliminating water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His404A | hydrogen bond acceptor |
| Lys353A | hydrogen bond donor, electrostatic stabiliser |
| Arg360A | hydrogen bond donor, electrostatic stabiliser |
| Arg490A | hydrogen bond donor, electrostatic stabiliser |
| Ser402A | hydrogen bond donor, electrostatic stabiliser |
| Gly403A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| His496A | covalently attached, metal ligand |
Chemical Components
ingold: acidic bimolecular nucleophilic substitution, intermediate collapse, intermediate formation, overall product formed, dehydration, coordination to a metal ion, decoordination from a metal ion
Step 3. One of the equatorial oxo groups deprotonates the attached hydrogen peroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys353A | hydrogen bond donor, electrostatic stabiliser |
| Arg360A | hydrogen bond donor, electrostatic stabiliser |
| Arg490A | hydrogen bond donor, electrostatic stabiliser |
| Ser402A | hydrogen bond donor, electrostatic stabiliser |
| Gly403A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| His496A | covalently attached, metal ligand |
Chemical Components
proton transfer, intermediate formation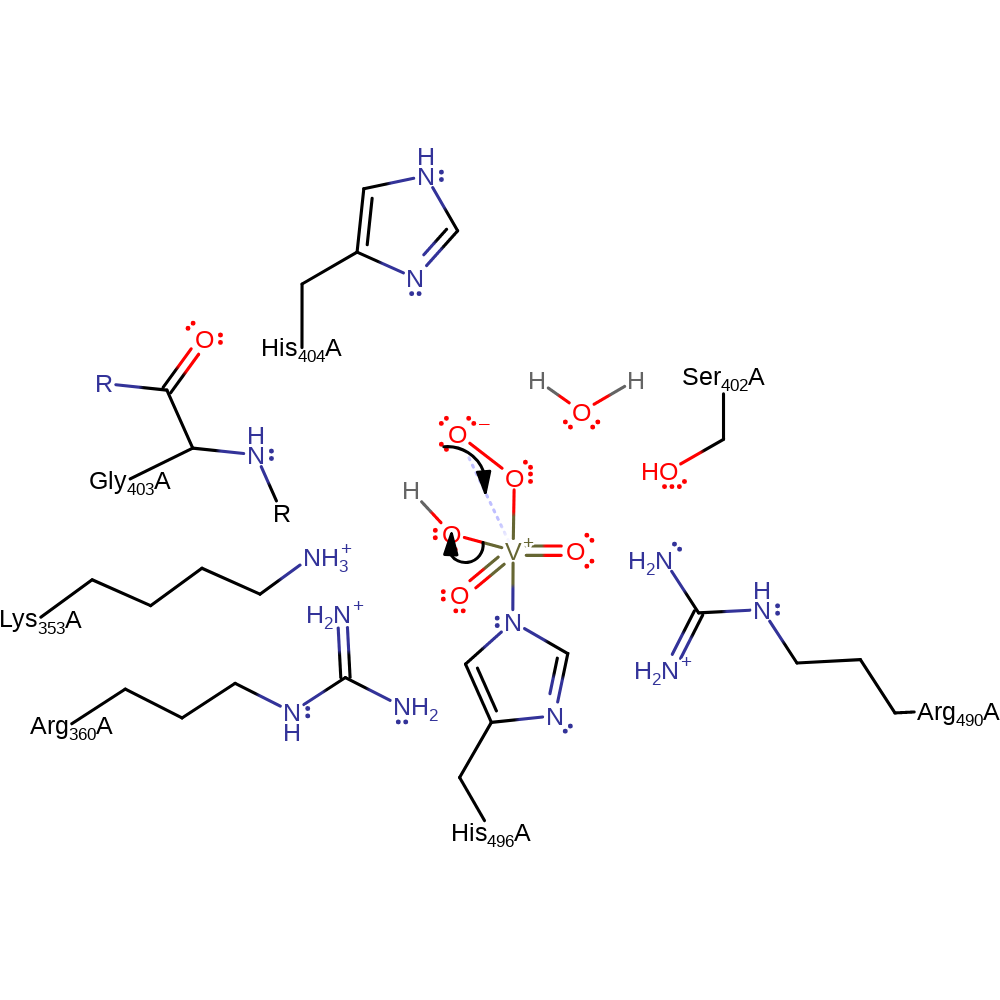
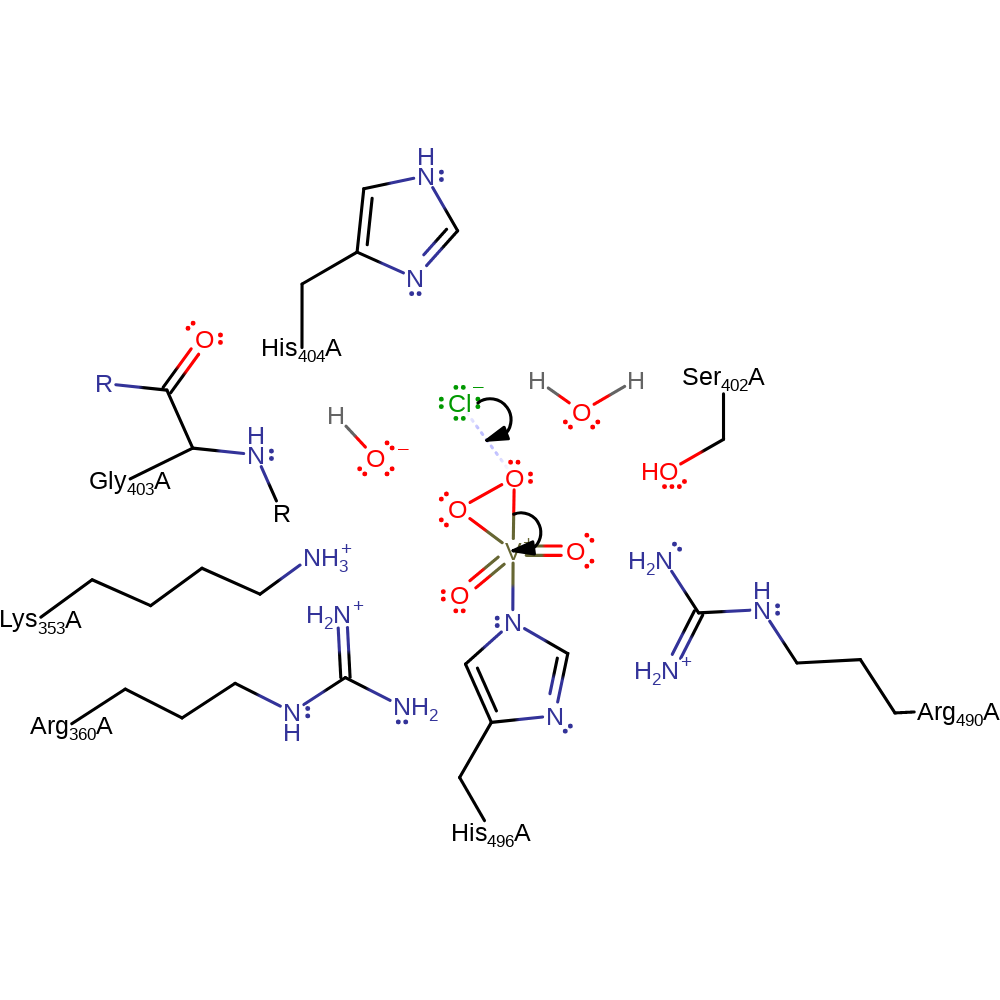
Step 4. The peroxide initiates a nucleophilic attack on the vanadate in a substitution reaction, eliminating hydroxide and forming a three membered ring.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys353A | hydrogen bond donor, electrostatic stabiliser |
| Arg360A | hydrogen bond donor, electrostatic stabiliser |
| Arg490A | hydrogen bond donor, electrostatic stabiliser |
| Ser402A | hydrogen bond donor, electrostatic stabiliser |
| Gly403A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| His496A | covalently attached, metal ligand |
Chemical Components
ingold: intramolecular nucleophilic substitution, intermediate formation, cyclisation, coordination to a metal ion, decoordination from a metal ionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys353A | hydrogen bond donor, electrostatic stabiliser |
| Arg360A | hydrogen bond donor, electrostatic stabiliser |
| Arg490A | hydrogen bond donor, electrostatic stabiliser |
| Ser402A | hydrogen bond donor, electrostatic stabiliser |
| Gly403A (main-N) | hydrogen bond donor |
| His496A | covalently attached, metal ligand |
Chemical Components
coordination, overall reactant used, intermediate formation
Step 6. The dioxygen bond undergoes rearrangement to eliminate hypochloroate with concomitant deprotonation of water, which initiates a nucleophilic attack on the vanadate in a coordination reaction to regenerate the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys353A | hydrogen bond donor, electrostatic stabiliser |
| Arg360A | hydrogen bond donor, electrostatic stabiliser |
| Arg490A | hydrogen bond donor, electrostatic stabiliser |
| Ser402A | hydrogen bond donor, electrostatic stabiliser |
| Gly403A (main-N) | hydrogen bond donor |
| His496A | covalently attached, metal ligand |





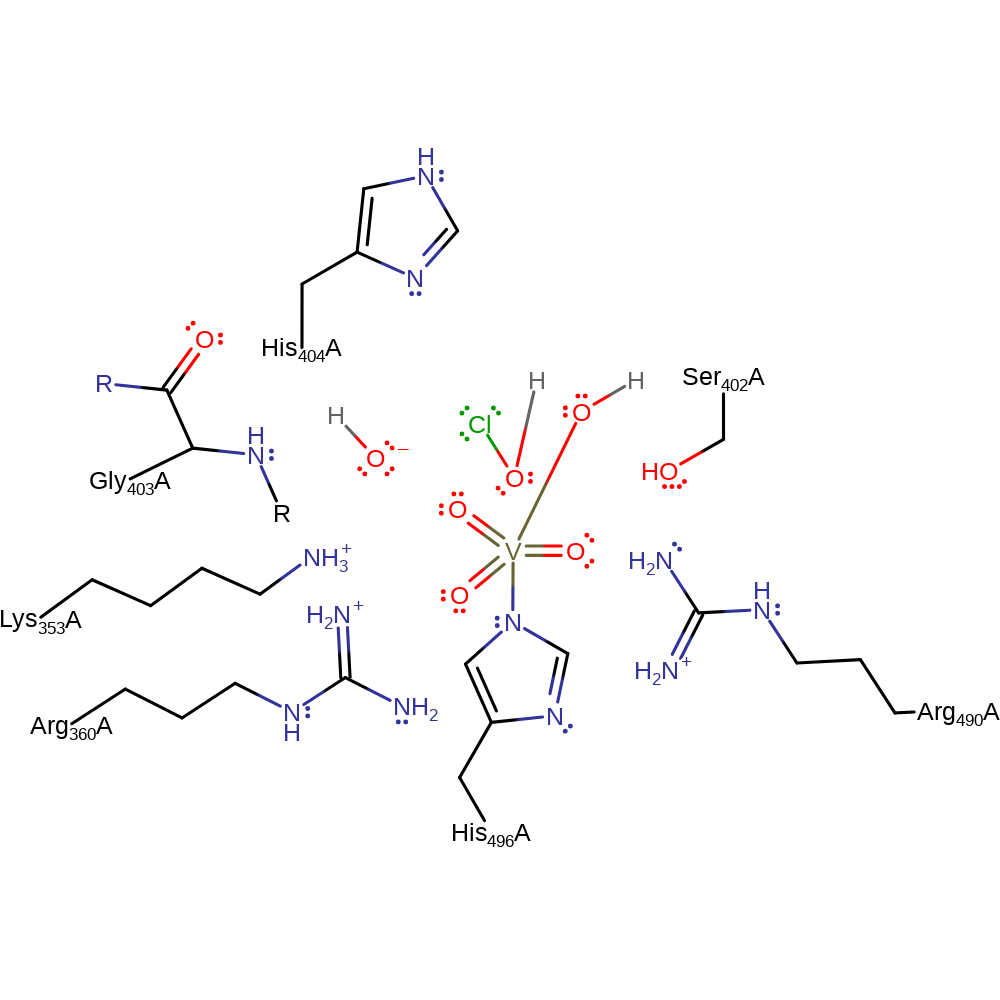 Download:
Download: