Cytochrome-c3 hydrogenase
Hydrogenases are enzymes that catalyse the reversible heterolytic dissociation of molecular hydrogen H2 to two protons and two electrons. The largest class of hydrogenases contains a NiFe center that is believed to be the catalytic site for hydrogen activation.The active site comprises a heterobimetallic cluster of Ni and Fe atoms. The bridging ligand X is proposed to be sulphur species due to the release of H2S during the course of the reaction.
Several alternative mechanisms exist for hydrogen activation by transition metal complexes. Generally these reactions proceed through either oxidative addition or sigma-bond metathesis (which is usually described as homolytic and without strong charge polarisation unless R is a polar group). Hydrogen isotope exchange and computational experiments for this enzyme indicate that the H-H cleavage reaction is heterolytic [PMID:11703120]. Cytochrome c3, the physiological electron acceptor for this particular hydrogenase, is a tetraheme cytochrome. The hemes all have different potentials, and probably only one or two are likely to accept electrons from the enzyme under physiological conditions.
Reference Protein and Structure
- Sequence
-
P12944
 (1.12.2.1)
(1.12.2.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Desulfovibrio gigas (Bacteria)

- PDB
-
2frv
- CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF NI-FE HYDROGENASE
(2.54 Å)



- Catalytic CATH Domains
-
1.10.645.10
 (see all for 2frv)
(see all for 2frv)
- Cofactors
- Tri-mu-sulfido-mu3-sulfido-triiron(0) (1), Tetra-mu3-sulfido-tetrairon (2), Carbonmonoxide-(dicyano) iron (1), Nickel(2+) (1) Metal MACiE
Enzyme Reaction (EC:1.12.2.1)
Enzyme Mechanism
Introduction
Dihydrogen coordinates to the Ni(II) centre, and a single electron is transferred from Ni(II) to the electron acceptor through the iron-sulfur clusters. Cys530 acts as a general base, deprotonating the hydrogen molecule, which forms a hydride ion that bridges the two metal centres. Cys530 then loses its proton to bulk solvent in which another residue (Glu18) acts as a proton relay. Cys530 accepts the bridging hydride, with the electron pair being donated to the Ni(III) centre. Cys530 then loses its proton to bulk solvent in which another residue (Glu18) acts as a proton relay. A single electron is transferred from the Ni(III) centre to the electron acceptor via the iron-sulfur clusters.
Catalytic Residues Roles
| UniProt | PDB* (2frv) | ||
| Cys530 | Cys530D(F) | Forms part of the nickel binding site, it also acts as a general acid/base during the course of the reaction. | hydrogen bond acceptor, hydrogen bond donor, metal ligand, proton acceptor, proton donor |
| Glu18 | Glu18D(F) | Acts as a general acid/base as part of the proton relay chain that removes the protons produced during the course of the reaction from the active site. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Cys65 | Cys65D(F) | Forms part of the nickel binding site. | activator, metal ligand |
| Cys533, Cys68 | Cys533D(F), Cys68D(F) | Act as bridging ligands between the iron and nickel active centre. | activator, metal ligand, electrostatic stabiliser |
| Arg463, Ser486, His72 | Arg463D(F), Ser486D(F), His72D(F) | Act to stabilise the electrostatic state of the NiFe centre. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
coordination, electron transfer, coordination to a metal ion, proton transfer, decoordination from a metal ion, proton relay, native state of enzyme regenerated, native state of cofactor regenerated, electron relayReferences
- Ash PA et al. (2017), ACS Catal, 7, 2471-2485. Proton Transfer in the Catalytic Cycle of [NiFe] Hydrogenases: Insight from Vibrational Spectroscopy. DOI:10.1021/acscatal.6b03182. PMID:28413691.
- Qiu S et al. (2016), Phys Chem Chem Phys, 18, 15369-15374. Why is a proton transformed into a hydride by [NiFe] hydrogenases? An intrinsic reactivity analysis based on conceptual DFT. DOI:10.1039/c6cp00948d. PMID:27210596.
- Greene BL et al. (2016), J Am Chem Soc, 138, 13013-13021. Glutamate Gated Proton-Coupled Electron Transfer Activity of a [NiFe]-Hydrogenase. DOI:10.1021/jacs.6b07789. PMID:27617712.
- Ogata H et al. (2016), J Biochem, 160, 251-258. Structure and function of [NiFe] hydrogenases. DOI:10.1093/jb/mvw048. PMID:27493211.
- Ogata H et al. (2015), Nature, 520, 571-574. Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase. DOI:10.1038/nature14110. PMID:25624102.
- Barilone JL et al. (2015), Phys Chem Chem Phys, 17, 16204-16212. Structural differences between the active sites of the Ni-A and Ni-B states of the [NiFe] hydrogenase: an approach by quantum chemistry and single crystal ENDOR spectroscopy. DOI:10.1039/c5cp01322d. PMID:26035632.
- Ogata H et al. (2015), Nat Commun, 6, 7890-. Hydride bridge in [NiFe]-hydrogenase observed by nuclear resonance vibrational spectroscopy. DOI:10.1038/ncomms8890. PMID:26259066.
- Murphy BJ et al. (2015), J Am Chem Soc, 137, 8484-8489. Discovery of Dark pH-Dependent H+Migration in a [NiFe]-Hydrogenase and Its Mechanistic Relevance: Mobilizing the Hydrido Ligand of the Ni-C Intermediate. DOI:10.1021/jacs.5b03182. PMID:26103582.
- Bruschi M et al. (2014), J Am Chem Soc, 136, 1803-1814. Disclosure of Key Stereoelectronic Factors for Efficient H2Binding and Cleavage in the Active Site of [NiFe]-Hydrogenases. DOI:10.1021/ja408511y. PMID:24392667.
- Rippers Y et al. (2012), Chemphyschem, 13, 3852-3856. Revealing the Absolute Configuration of the CO and CN−Ligands at the Active Site of a [NiFe] Hydrogenase. DOI:10.1002/cphc.201200562. PMID:22945586.
- Kampa M et al. (2012), J Biol Inorg Chem, 17, 1269-1281. Computational study of the electronic structure and magnetic properties of the Ni–C state in [NiFe] hydrogenases including the second coordination sphere. DOI:10.1007/s00775-012-0941-9. PMID:23053531.
- Fritsch J et al. (2011), Biochemistry, 50, 5858-5869. [NiFe] and [FeS] Cofactors in the Membrane-Bound Hydrogenase ofRalstonia eutrophaInvestigated by X-ray Absorption Spectroscopy: Insights into O2-Tolerant H2Cleavage. DOI:10.1021/bi200367u. PMID:21618994.
- Pandelia ME et al. (2010), Chemphyschem, 11, 1127-1140. Intermediates in the Catalytic Cycle of [NiFe] Hydrogenase: Functional Spectroscopy of the Active Site. DOI:10.1002/cphc.200900950. PMID:20301175.
- Jayapal P et al. (2008), Phys Chem Chem Phys, 10, 4249-4257. QM/MM studies of Ni–Fe hydrogenases: the effect of enzyme environment on the structure and energies of the inactive and active states. DOI:10.1039/b804035d. PMID:18633545.
- De Lacey AL et al. (2007), Chem Rev, 107, 4304-4330. Activation and Inactivation of Hydrogenase Function and the Catalytic Cycle: Spectroelectrochemical Studies. DOI:10.1021/cr0501947. PMID:17715982.
- Pardo A et al. (2006), J Biol Inorg Chem, 11, 286-306. Density functional study of the catalytic cycle of nickel-iron [NiFe] hydrogenases and the involvement of high-spin nickel(II). DOI:10.1007/s00775-005-0076-3. PMID:16511689.
- Stein M et al. (2004), J Inorg Biochem, 98, 862-877. Relativistic DFT calculation of the reaction cycle intermediates of [NiFe] hydrogenase: a contribution to understanding the enzymatic mechanism☆. DOI:10.1016/j.jinorgbio.2004.03.002. PMID:15134933.
- M. Stein (2001), PhD thesis, Technische Unversitat Berlin, 1, 171-183. Insight into the Mechanism of [NiFe] Hydrogenase by means of Magnetic Resonance Experiments and DFT Calculation. PMID:Stein2001.
- Niu S et al. (2001), Inorg Chem, 40, 6201-6203. Modeling the Active Sites in Metalloenzymes 5. The Heterolytic Bond Cleavage of H2in the [NiFe] Hydrogenase ofDesulfovibrio gigasby a Nucleophilic Addition Mechanism. DOI:10.1021/ic0107274. PMID:11703120.
- Stein M et al. (2001), J Am Chem Soc, 123, 5839-5840. Relativistic DFT Calculations of the Paramagnetic Intermediates of [NiFe] Hydrogenase. Implications for the Enzymatic Mechanism. DOI:10.1021/ja005808y. PMID:11403633.
- Higuchi Y et al. (1999), Structure, 7, 549-556. Removal of the bridging ligand atom at the Ni–Fe active site of [NiFe] hydrogenase upon reduction with H2, as revealed by X-ray structure analysis at 1.4 Å resolution. DOI:10.1016/s0969-2126(99)80071-9. PMID:10378274.
- Volbeda A et al. (1995), Nature, 373, 580-587. Crystal structure of the nickel–iron hydrogenase from Desulfovibrio gigas . DOI:10.1038/373580a0. PMID:7854413.
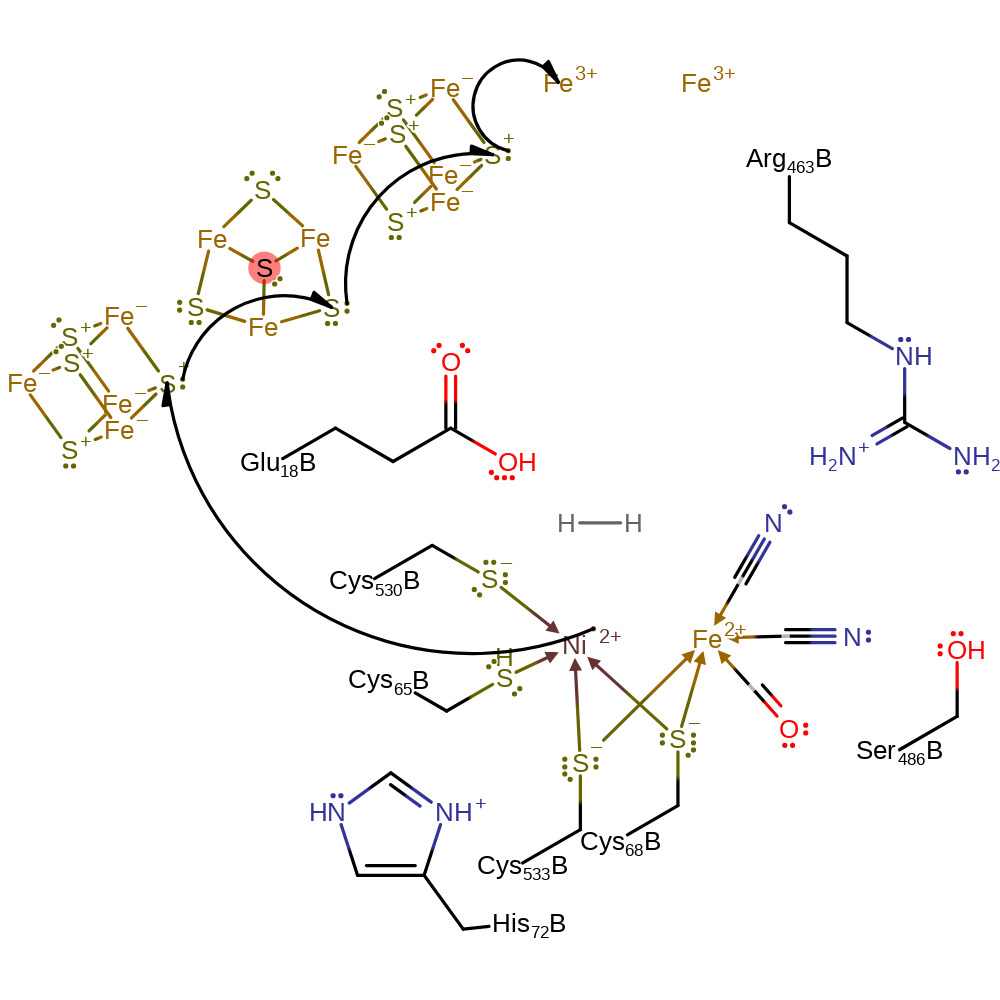
Step 1. Hydrogen molecules diffuse from the surface of the protein and this is unlikely to be rate-limiting. Hydrogen easily permeates the protein and moves preferentially in channels to the Ni side of the active site. [PMID:17715982]. Dihydrogen coordinates to the Ni(II) centre, and a single electron is transferred from Ni(II) to the electron acceptor through the iron-sulfur clusters.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys68D(F) | metal ligand |
| Cys533D(F) | metal ligand |
| Cys530D(F) | metal ligand |
| Cys65D(F) | metal ligand |
| Arg463D(F) | hydrogen bond donor, electrostatic stabiliser |
| Ser486D(F) | hydrogen bond donor, electrostatic stabiliser |
| Cys65D(F) | activator |
| Cys533D(F) | activator, electrostatic stabiliser |
| Cys68D(F) | activator, electrostatic stabiliser |
| His72D(F) | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
coordination, electron transfer, coordination to a metal ion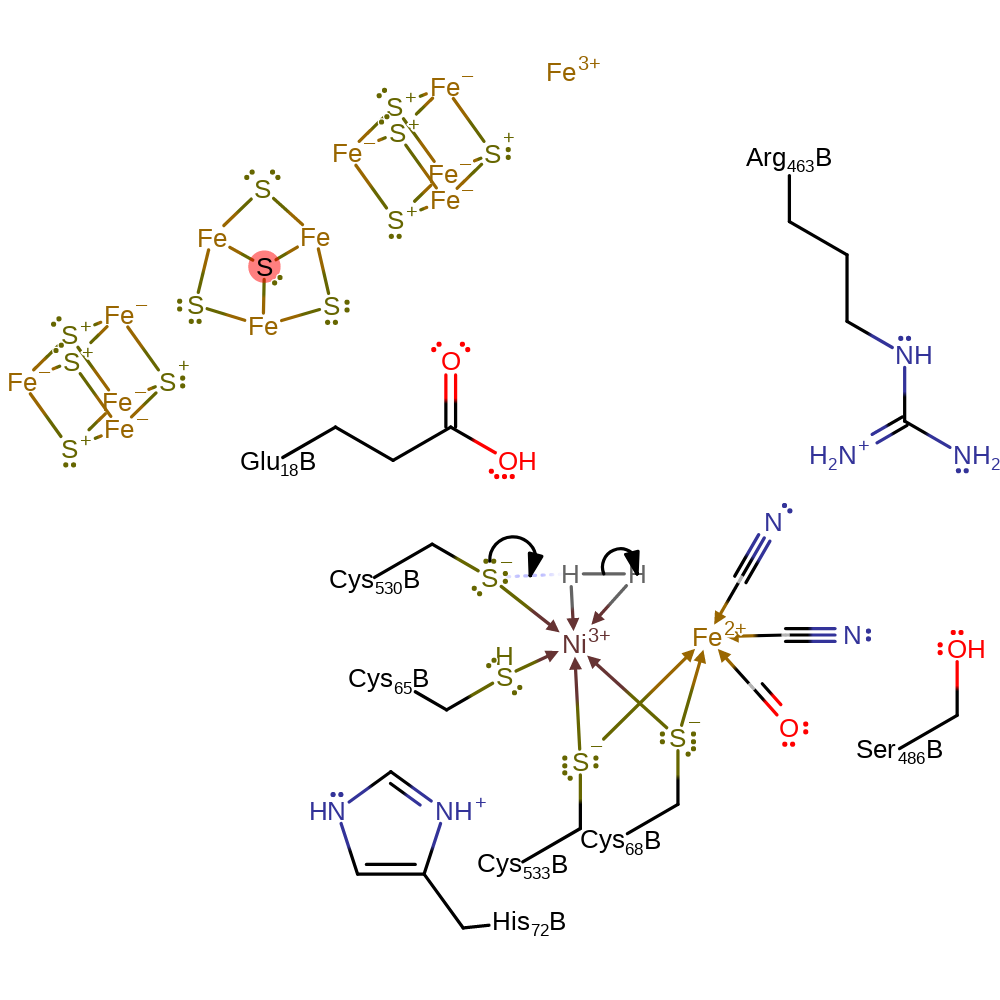
Step 2. Cys530 acts as a general base, deprotonating the hydrogen molecules, which forms a hydride ion that bridges the two metal centres.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys68D(F) | metal ligand |
| Cys533D(F) | metal ligand |
| Cys530D(F) | metal ligand |
| Cys65D(F) | metal ligand |
| Arg463D(F) | hydrogen bond donor, electrostatic stabiliser |
| Ser486D(F) | hydrogen bond donor, electrostatic stabiliser |
| Cys530D(F) | hydrogen bond acceptor |
| Cys65D(F) | activator |
| Cys533D(F) | activator, electrostatic stabiliser |
| Cys68D(F) | activator, electrostatic stabiliser |
| His72D(F) | hydrogen bond donor, electrostatic stabiliser |
| Cys530D(F) | proton acceptor |
Chemical Components
coordination, proton transfer, coordination to a metal ion, decoordination from a metal ion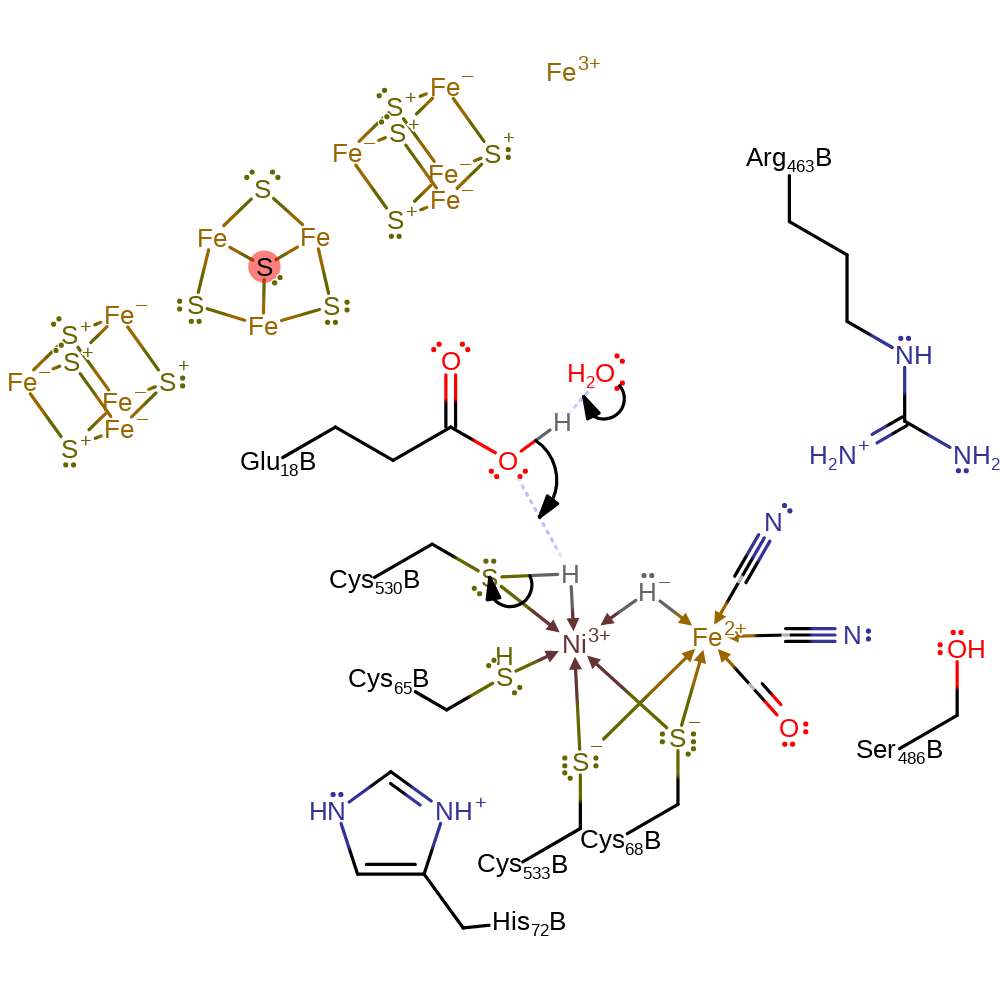
Step 3. Cys530 loses its proton to bulk solvent in which Glu18 acts as a proton relay.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys68D(F) | metal ligand |
| Cys533D(F) | metal ligand |
| Cys530D(F) | metal ligand |
| Cys65D(F) | metal ligand |
| Arg463D(F) | hydrogen bond donor, electrostatic stabiliser |
| Ser486D(F) | hydrogen bond donor, electrostatic stabiliser |
| Cys530D(F) | hydrogen bond donor |
| Cys65D(F) | activator |
| Cys533D(F) | activator, electrostatic stabiliser |
| Cys68D(F) | activator, electrostatic stabiliser |
| His72D(F) | hydrogen bond donor, electrostatic stabiliser |
| Glu18D(F) | hydrogen bond acceptor, hydrogen bond donor, proton relay, proton acceptor, proton donor |
| Cys530D(F) | proton donor |
Chemical Components
proton transfer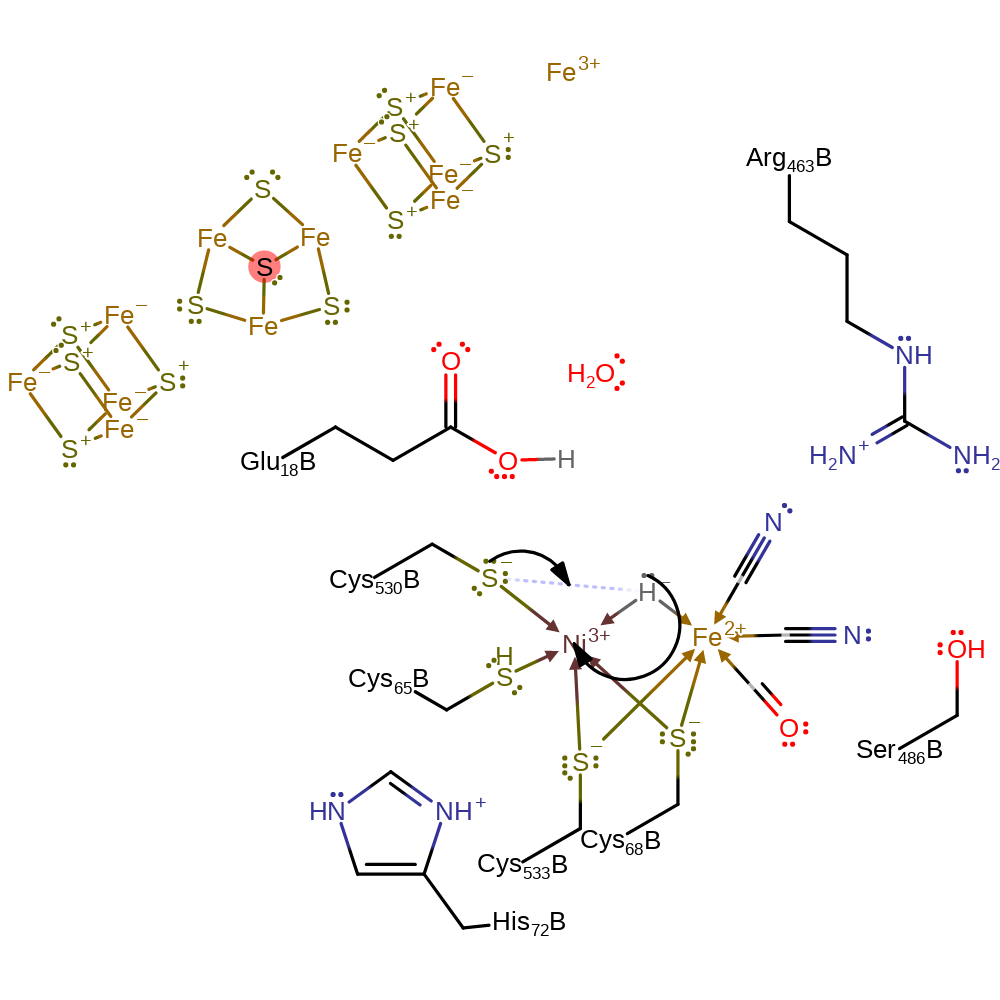
Step 4. Cys530 acts as a general base, accepting the bridging hydride, with the electron pair being donated to the Ni(III) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys68D(F) | metal ligand |
| Cys533D(F) | metal ligand |
| Cys530D(F) | metal ligand |
| Cys65D(F) | metal ligand |
| Arg463D(F) | hydrogen bond donor, electrostatic stabiliser |
| Ser486D(F) | hydrogen bond donor, electrostatic stabiliser |
| Cys530D(F) | hydrogen bond acceptor |
| Cys65D(F) | activator |
| Cys533D(F) | activator, electrostatic stabiliser |
| Cys68D(F) | activator, electrostatic stabiliser |
| His72D(F) | hydrogen bond donor, electrostatic stabiliser |
| Cys530D(F) | proton acceptor |
Chemical Components
proton transfer, decoordination from a metal ion, electron transfer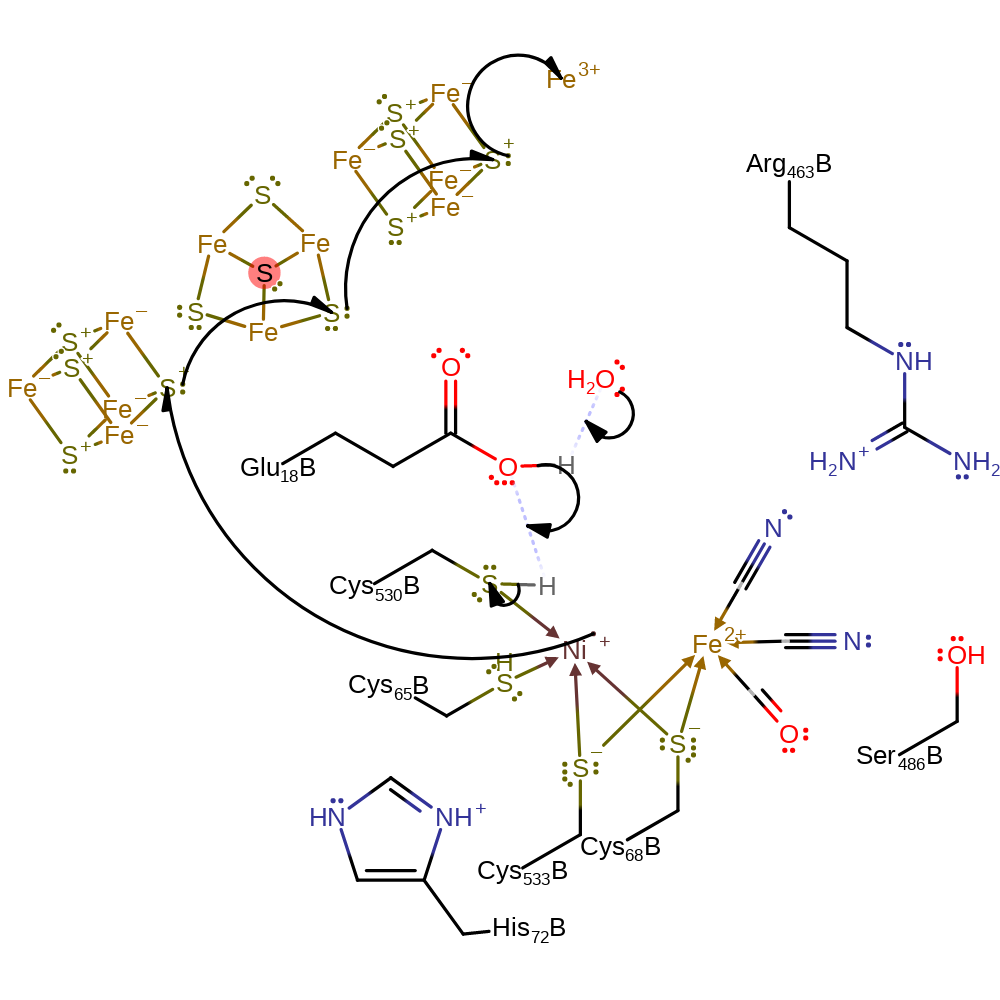
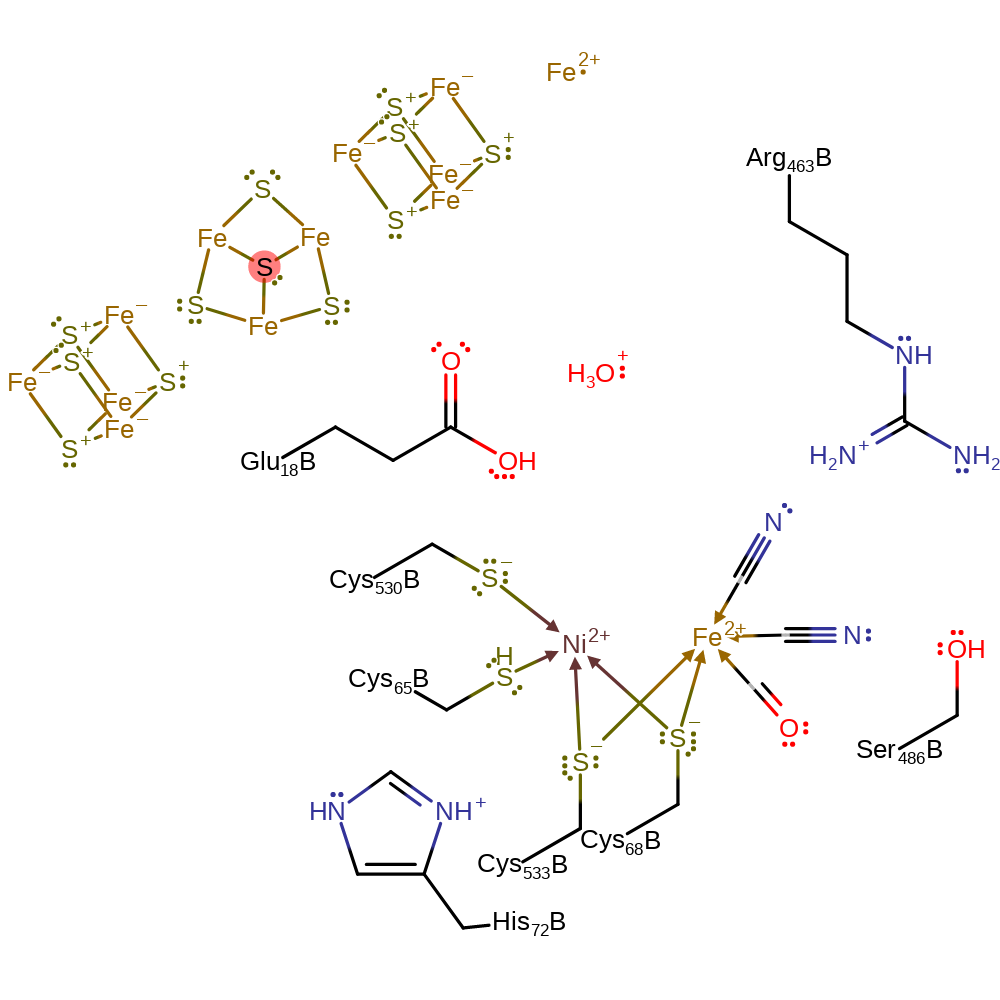
Step 5. Cys530 loses its proton to bulk solvent in which Glu18 acts as a proton relay. A single electron is transferred from the Ni(III) centre to the electron acceptor via the iron-sulfur clusters.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg463D(F) | hydrogen bond donor, electrostatic stabiliser |
| Ser486D(F) | hydrogen bond donor, electrostatic stabiliser |
| Cys530D(F) | hydrogen bond donor |
| Cys65D(F) | activator |
| Cys533D(F) | activator, electrostatic stabiliser |
| Cys68D(F) | activator, electrostatic stabiliser |
| His72D(F) | hydrogen bond donor, electrostatic stabiliser |
| Glu18D(F) | hydrogen bond acceptor, hydrogen bond donor, proton relay |
| Cys68D(F) | metal ligand |
| Cys533D(F) | metal ligand |
| Cys530D(F) | metal ligand |
| Cys65D(F) | metal ligand |
| Glu18D(F) | proton donor, proton acceptor |
| Cys530D(F) | proton donor |




 Download:
Download: