Pyruvate dehydrogenase (acetyl-transferring)
Pyruvate decarboxylase (E1) catalyzes the first two reactions of the four involved in oxidative decarboxylation of pyruvate by the pyruvate dehydrogenase (PDH) multienzyme complex. It requires thiamin diphosphate to bring about the decarboxylation of pyruvate, which is followed by the reductive acetylation of a lipoyl group covalently bound to the N(6) amino group of a lysine residue in the second catalytic component, a dihydrolipoyl acetyltransferase (E2).
The lipoamide is covalently bound to dihydrolipoamide acetyltransferase (the second catalytic component of the Pyruvate Dehydrogenase Complex) forming (dihydrolipoyllysine-residue acetyltransferase) lipoyllysine [PMID:12795594]. The enzyme consists of two catalytic sites, each providing TDP (thiamine diphosphate) and magnesium as cofactors and each formed on the interface between the aplha and beta subunits [PMID:12651851].
Reference Protein and Structure
- Sequences
-
P21874
 (1.2.4.1)
(1.2.4.1)
P21873 (1.2.4.1)
(1.2.4.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Geobacillus stearothermophilus (Bacteria)

- PDB
-
1w85
- The crystal structure of pyruvate dehydrogenase E1 bound to the peripheral subunit binding domain of E2
(2.0 Å)



- Catalytic CATH Domains
-
3.40.50.970
 (see all for 1w85)
(see all for 1w85)
- Cofactors
- Magnesium(2+) (1), Thiamine(1+) diphosphate(3-) (1) Metal MACiE
Enzyme Reaction (EC:1.2.4.1)
Enzyme Mechanism
Introduction
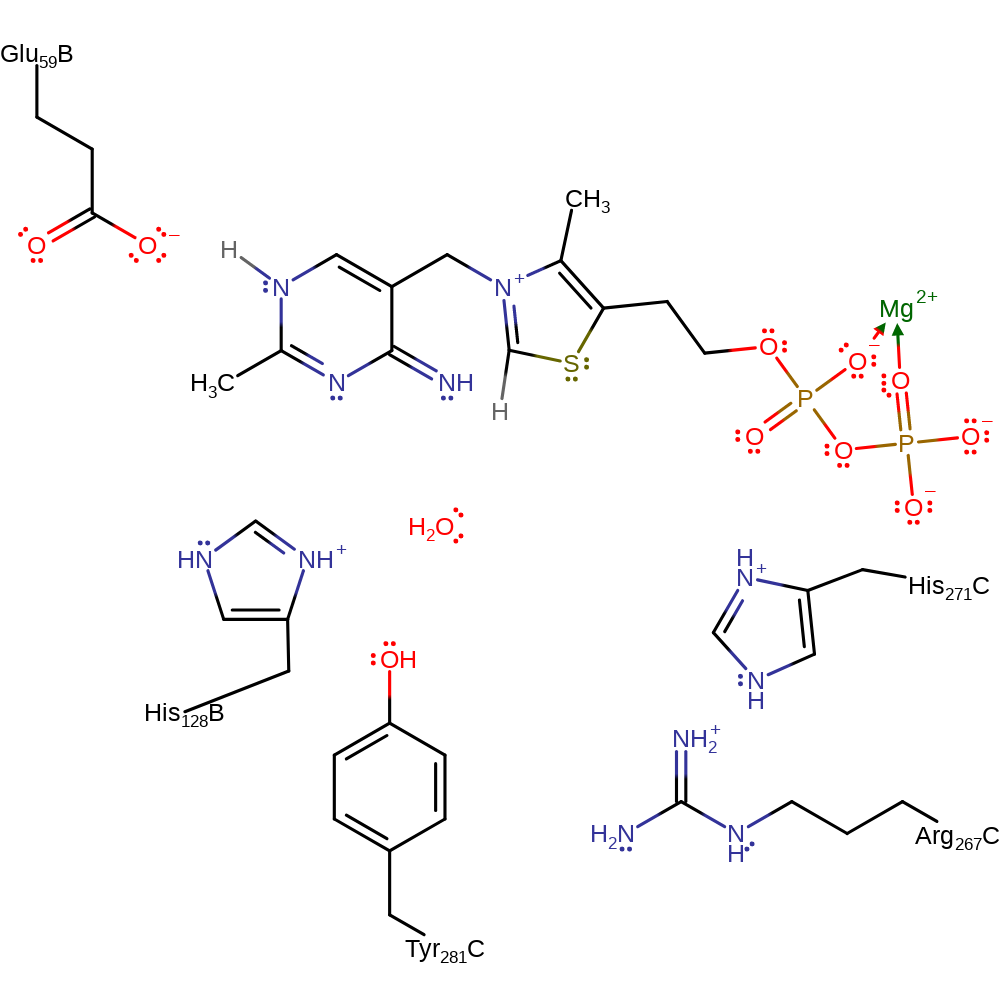
The TPP (thiamine diphosphate) cofactor is activated by the formation of an ionic N1'-H...O bond between the N1' atom of the aminopyrimidine ring of the cofactor and the OG of the intrinsic Glu59, resulting in the transfer of a negative charge from the protein to the cofactor. This transfer then imposes an active V-conformation that brings the N4' atom of the cofactor to the distance required for the intramolecular C2-H...N4' hydrogen bond with the thiazolium C2 atom. This initial activation is then followed by abstraction of the proton from the C2 atom. The resulting carbanion of thiamine diphosphate initiates a nucleophilic attack on the carbonyl carbon of pyruvate in an addition reaction. The puruvate is now covalently attached to the activated TPP. His271C donates a proton to the newly formed oxyanion. The covalently bound pyruvate undergoes decarboxylation and the carbanion formed initiates a nucleophilic attack on the second substrate (lipoamide) in an addition reaction with concomitant deprotonation of His128. Lipoamide is covalently bound to dihydrolipoamide acetyltransferase (the second catalytic component of the Pyruvate Dehydrogenase Complex) forming [dihydrolipoyllysine-residue acetyltransferase] lipoyllysine. His271C deprotonates the hydroxyl in an elimination which reforms the carbanionic form of ThDP and produces the N6-[(R)-S8-acetyldihydrolipoyl]-L-lysine residue product. Finally, the carbanion of the thiamine diphosphate cofactor deprotonates the adjacent amine, which initiates double bond rearrangement that results in the deprotonation of Glu59 and a bulk solvent molecule retruns His128 to its starting protonation state.
Catalytic Residues Roles
| UniProt | PDB* (1w85) | ||
| Glu60 | Glu59B | Acts as a general acid/base in the activation of the thiamine diphosphate cofactor. | proton acceptor, proton donor |
| His129, His272 | His128B, His271C | Acts as a general acid/base. | proton acceptor, proton donor |
| Arg268, Tyr282 | Arg267C, Tyr281C | Help stabilise the reactive intermediates formed. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, cofactor used, intermediate formation, bimolecular nucleophilic addition, overall reactant used, unimolecular elimination by the conjugate base, decarboxylation, overall product formed, bimolecular elimination, intermediate collapse, native state of cofactor regenerated, intermediate terminated, native state of enzyme regenerated, inferred reaction stepReferences
- Fries M et al. (2003), Biochemistry, 42, 6996-7002. Reaction Mechanism of the Heterotetrameric (α2β2) E1 Component of 2-Oxo Acid Dehydrogenase Multienzyme Complexes†. DOI:10.1021/bi027397z. PMID:12795594.
- Sheng X et al. (2013), Biochemistry, 52, 8079-8093. Theoretical Study of the Catalytic Mechanism of E1 Subunit of Pyruvate Dehydrogenase Multienzyme Complex fromBacillus stearothermophilus. DOI:10.1021/bi400577f. PMID:24171427.
- Ciszak EM et al. (2003), J Biol Chem, 278, 21240-21246. Structural Basis for Flip-Flop Action of Thiamin Pyrophosphate-dependent Enzymes Revealed by Human Pyruvate Dehydrogenase. DOI:10.1074/jbc.m300339200. PMID:12651851.
- Aevarsson A et al. (1999), Nat Struct Biol, 6, 785-792. Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes. DOI:10.1038/11563. PMID:10426958.
- Schellenberger A (1998), Biochim Biophys Acta, 1385, 177-186. Sixty years of thiamin diphosphate biochemistry. DOI:10.1016/s0167-4838(98)00067-3. PMID:9655906.
- Hübner G et al. (1998), Biochim Biophys Acta, 1385, 221-228. Activation of thiamin diphosphate in enzymes. DOI:10.1016/s0167-4838(98)00070-3. PMID:9655909.
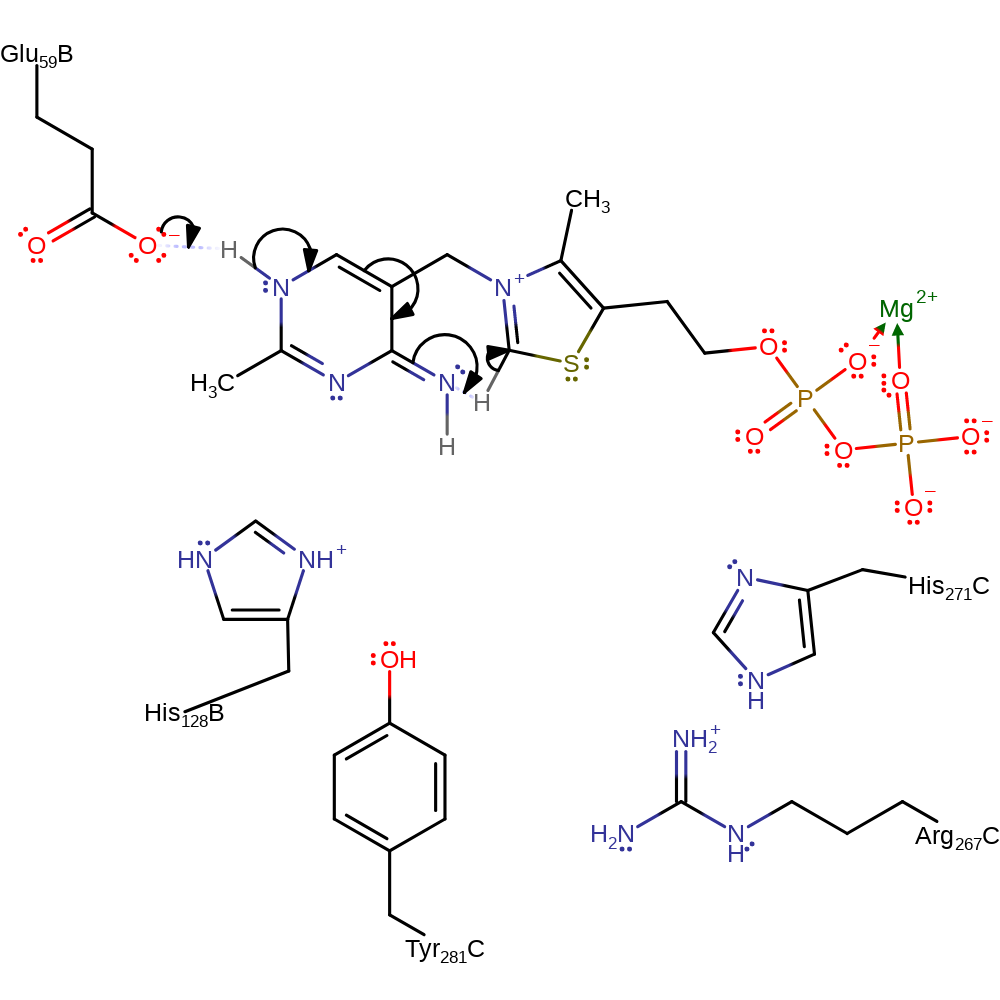
Step 1. Glu59 deprotonates the thiamine diphosphate cofactor, which initiates double bond rearrangement that results in the deprotonation of the N=CH-S group, activating the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu59B | proton acceptor |
Chemical Components
proton transfer, cofactor used, intermediate formation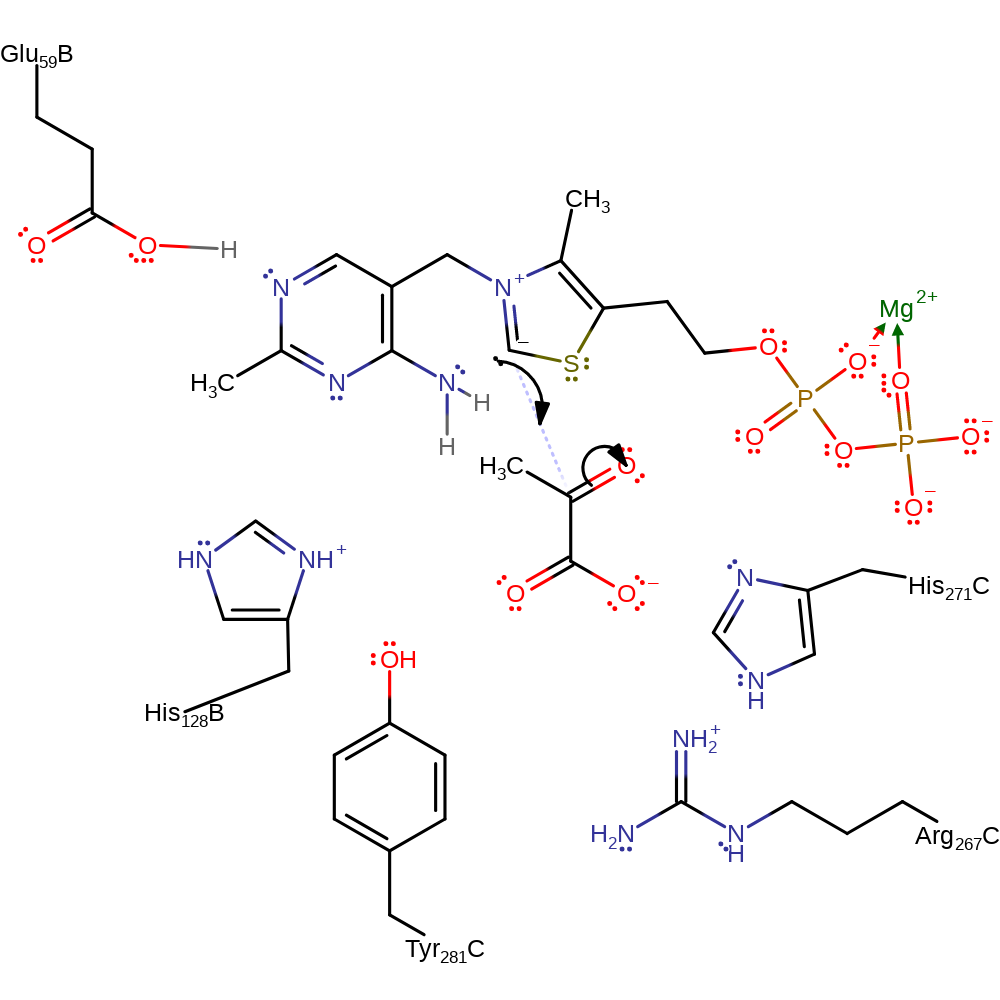
Step 2. The carbanion of thiamine diphosphate initiates a nucleophilic attack on the carbonyl carbon of pyruvate in an addition reaction
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg267C | hydrogen bond donor, electrostatic stabiliser |
| Tyr281C | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg267C | hydrogen bond donor, electrostatic stabiliser |
| His271C | hydrogen bond donor, electrostatic stabiliser |
| Tyr281C | hydrogen bond donor, electrostatic stabiliser |
| His271C | proton donor |
Chemical Components
proton transfer, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg267C | hydrogen bond donor, electrostatic stabiliser |
| His271C | hydrogen bond acceptor, electrostatic stabiliser |
| Tyr281C | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: unimolecular elimination by the conjugate base, decarboxylation, overall product formed, intermediate formation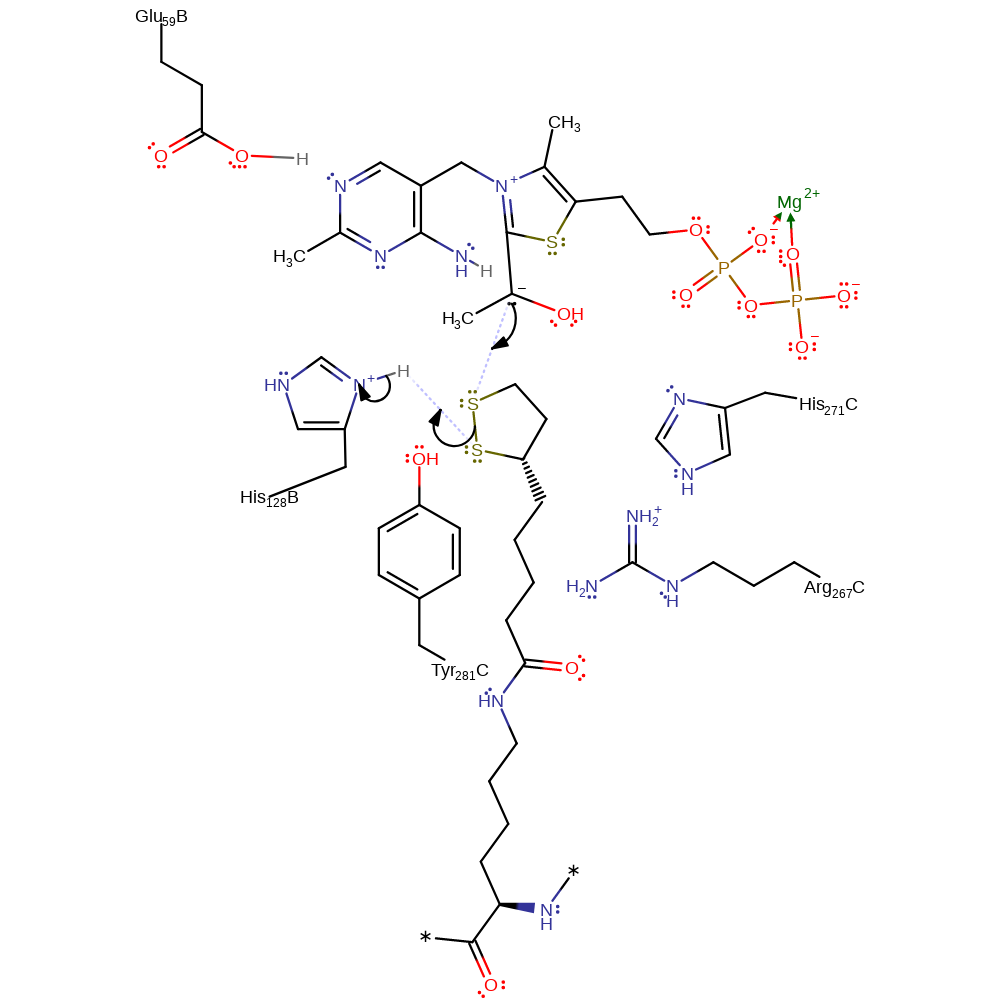
Step 5. The carbanion formed by decarboxylation initiates a nucleophilic attack on the second substrate in an addition reaction with concomitant deprotonation of His128.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His271C | hydrogen bond acceptor |
| His128B | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, intermediate formation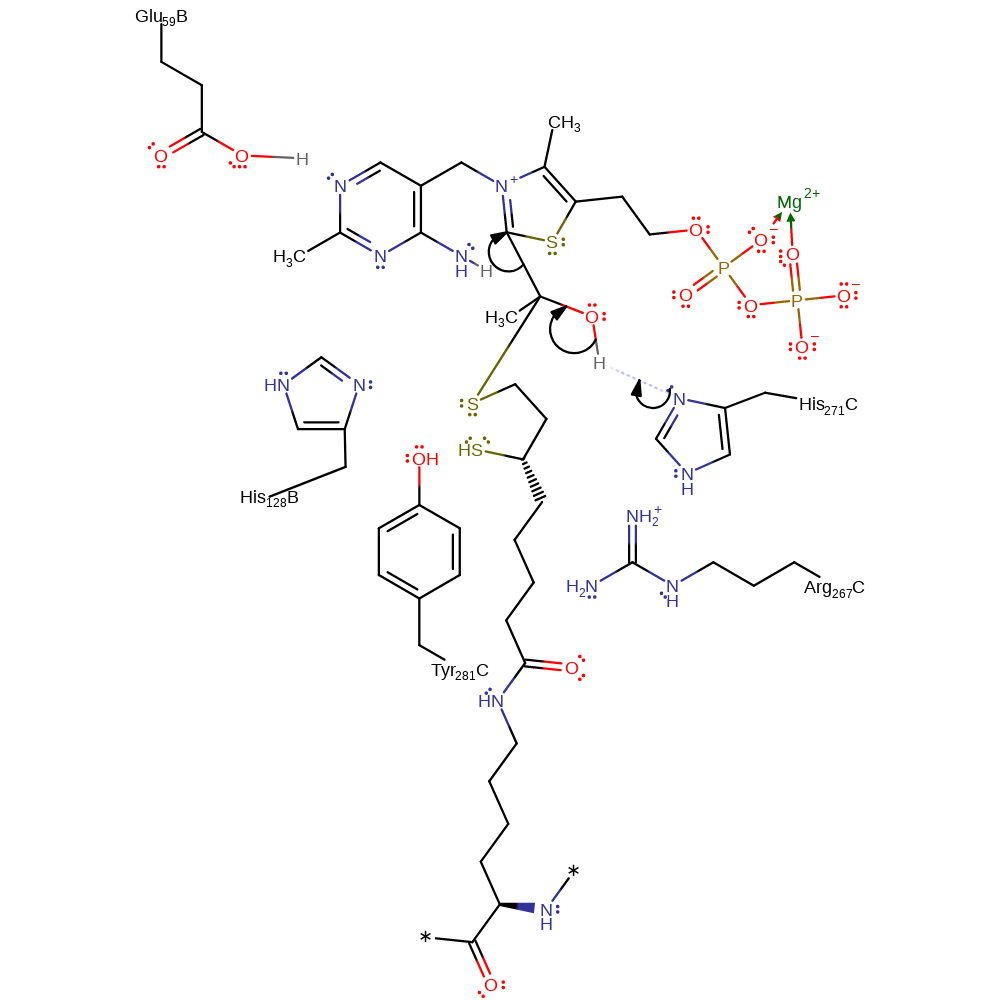
Step 6. His271C deprotonates the hydroxyl in an elimination which reforms the carbanionic form of ThDP and produces the second product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His271C | hydrogen bond acceptor |
| His271C | proton acceptor |
Chemical Components
ingold: bimolecular elimination, intermediate collapse, intermediate formation, overall product formed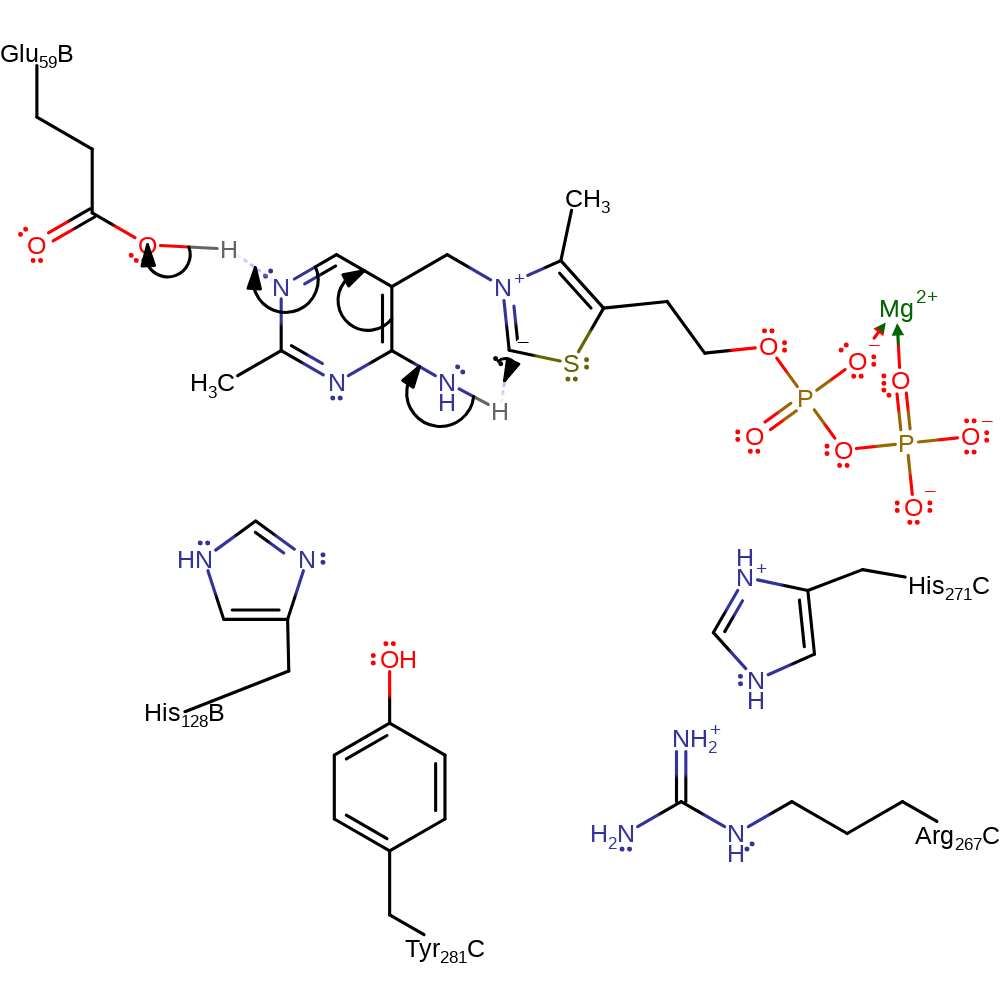
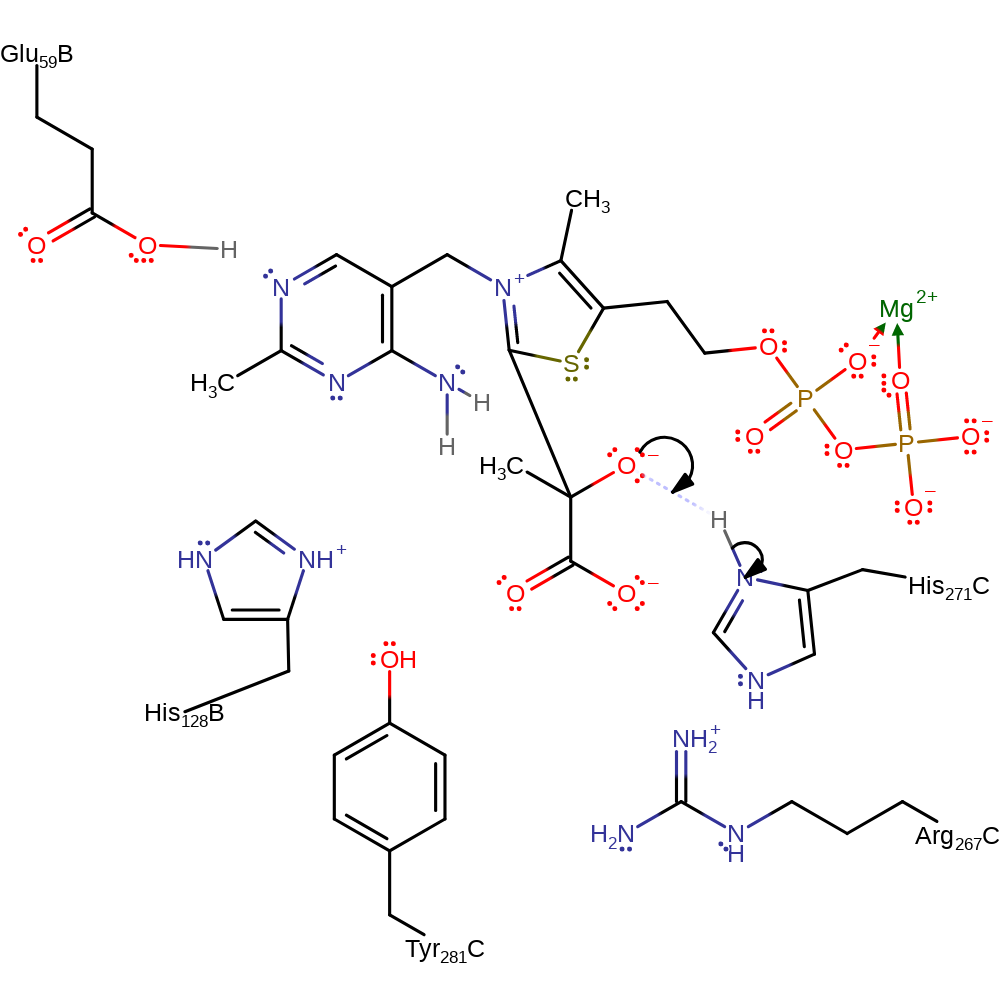
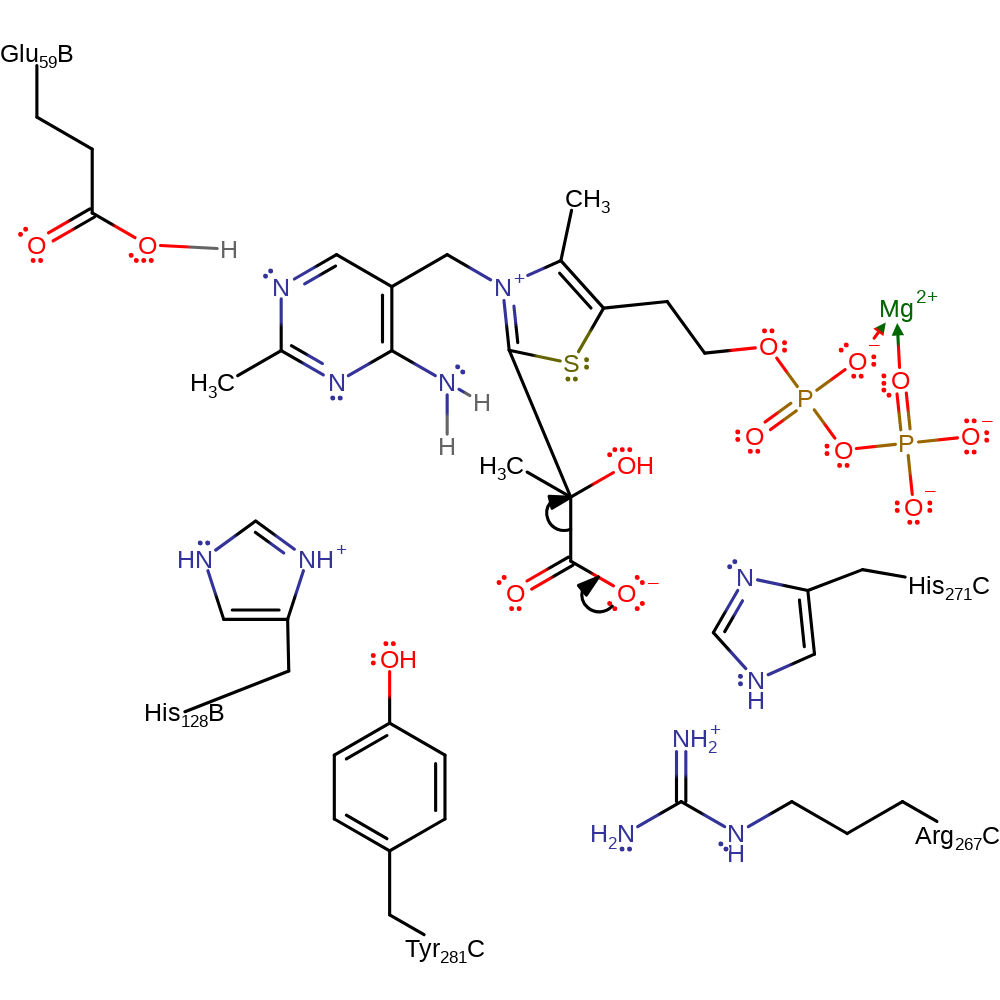
Step 7. The carbanion of the thiamine diphosphate cofactor deprotonates the adjacent amine, which initiates double bond rearrangement that results in the deprotonation of Glu59.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu59B | proton donor |
Chemical Components
proton transfer, native state of cofactor regenerated, intermediate terminatedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His128B | proton acceptor |





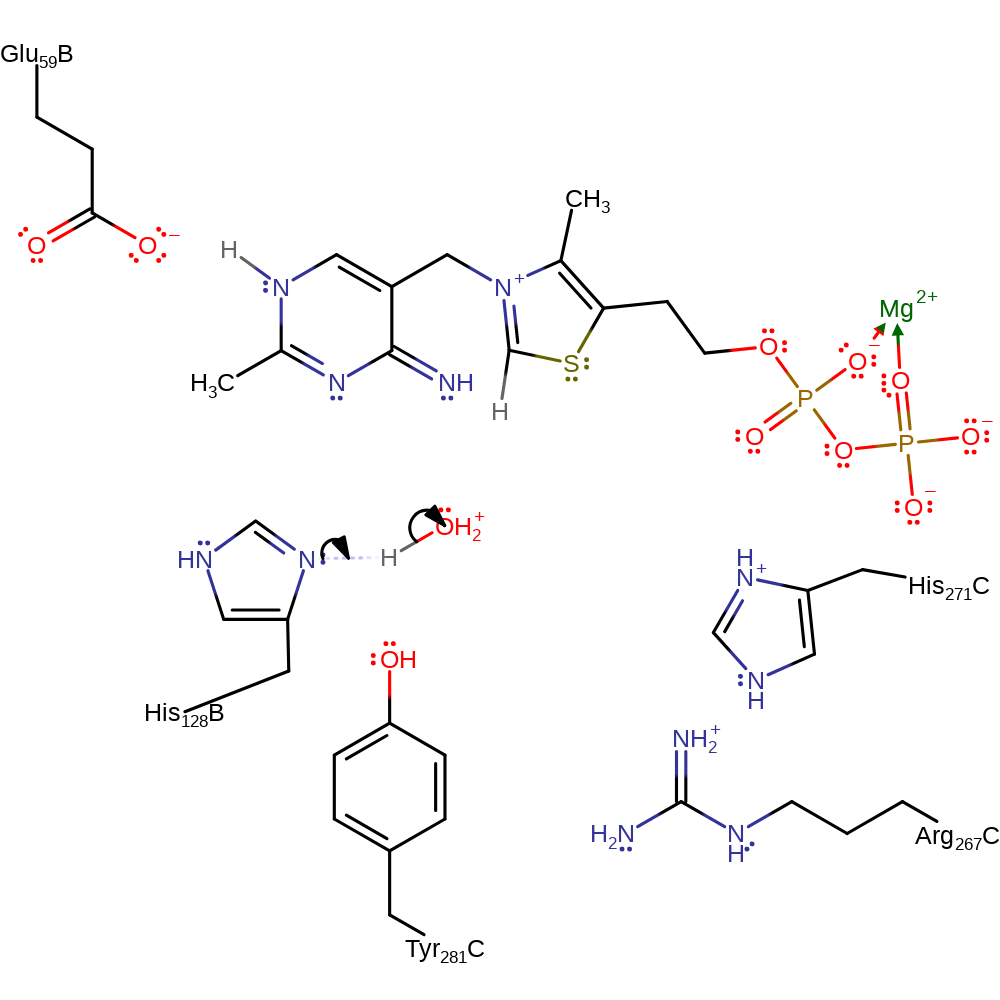
 Download:
Download: