 |
PDBsum entry 6giq

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Electron transport
|
PDB id
|
|

|

|
6giq
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 431 a.a.
431 a.a.
|
 |

|

|

|

|

|

|

|

 352 a.a.
352 a.a.
|
 |

|

|

|

|

|

|

|

 385 a.a.
385 a.a.
|
 |

|

|

|

|

|

|

|

 248 a.a.
248 a.a.
|
 |

|

|

|

|

|

|

|

 185 a.a.
185 a.a.
|
 |

|

|

|

|

|

|

|

 74 a.a.
74 a.a.
|
 |

|

|

|

|

|

|

|

 126 a.a.
126 a.a.
|
 |

|

|

|

|

|

|

|

 93 a.a.
93 a.a.
|
 |

|

|

|

|

|

|

|
 54 a.a.
54 a.a.
|
 |

|

|

|

|

|

|

|
 54 a.a.
54 a.a.
|
 |

|

|

|

|

|

|

|
 44 a.a.
44 a.a.
|
 |

|

|

|

|

|

|

|
 51 a.a.
51 a.a.
|
 |

|

|

|

|

|

|

|
 522 a.a.
522 a.a.
|
 |

|

|

|

|

|

|

|
 235 a.a.
235 a.a.
|
 |

|

|

|

|

|

|

|
 262 a.a.
262 a.a.
|
 |

|

|

|

|

|

|

|
 49 a.a.
49 a.a.
|
 |

|

|

|

|

|

|

|
 96 a.a.
96 a.a.
|
 |

|

|

|

|

|

|

|
 99 a.a.
99 a.a.
|
 |

|

|

|

|

|

|

|
 29 a.a.
29 a.a.
|
 |

|

|

|

|

|

|

|
 31 a.a.
31 a.a.
|
 |

|

|

|

|

|

|

|
 49 a.a.
49 a.a.
|
 |

|

|

|

|

|

|

|
 57 a.a.
57 a.a.
|
 |

|

|

|

|

|

|

|
 18 a.a.
18 a.a.
|
 |

|

|

|

|

|

|

|
 31 a.a.
31 a.a.
|
 |

|

|
|
|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Electron transport
|
 |
|
Title:
|
 |
Saccharomyces cerevisiae respiratory supercomplex iii2iv
|
|
Structure:
|
 |
Bj4_g0001550.mRNA.1.Cds.1. Chain: a, l. Synonym: cor1 isoform 1,cytochrome b-c1 complex subunit 1, mitochondrial,y55_g0001550.mRNA.1.Cds.1. Cytochrome b-c1 complex subunit 2, mitochondrial. Chain: b, m. Synonym: qcr2 isoform 1. Cytochrome b. Chain: c, n.
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Saccharomyces cerevisiae (strain atcc 204508 / s288c). Organism_taxid: 559292. Strain: atcc 204508 / s288c. Organism_taxid: 4932
|
|
Authors:
|
 |
S.Rathore,J.Berndtsson,J.Conrad,M.Ott
|
|
Key ref:
|
 |
S.Rathore
et al.
(2019).
Cryo-EM structure of the yeast respiratory supercomplex.
Nat Struct Mol Biol,
26,
50-57.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
15-May-18
|
Release date:
|
02-Jan-19
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07256
(QCR1_YEAST) -
Cytochrome b-c1 complex subunit 1, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
457 a.a.
431 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P07257
(QCR2_YEAST) -
Cytochrome b-c1 complex subunit 2, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
368 a.a.
352 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00163
(CYB_YEAST) -
Cytochrome b from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
385 a.a.
385 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P07143
(CY1_YEAST) -
Cytochrome c1, heme protein, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
309 a.a.
248 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P08067
(UCRI_YEAST) -
Cytochrome b-c1 complex subunit Rieske, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
215 a.a.
185 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00127
(QCR6_YEAST) -
Cytochrome b-c1 complex subunit 6, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
147 a.a.
74 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00128
(QCR7_YEAST) -
Cytochrome b-c1 complex subunit 7, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
127 a.a.
126 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P08525
(QCR8_YEAST) -
Cytochrome b-c1 complex subunit 8, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
94 a.a.
93 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P22289
(QCR9_YEAST) -
Cytochrome b-c1 complex subunit 9, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
66 a.a.
54 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P22289
(QCR9_YEAST) -
Cytochrome b-c1 complex subunit 9, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
66 a.a.
54 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P37299
(QCR10_YEAST) -
Cytochrome b-c1 complex subunit 10, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
77 a.a.
44 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P37299
(QCR10_YEAST) -
Cytochrome b-c1 complex subunit 10, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
77 a.a.
51 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00401
(COX1_YEAST) -
Cytochrome c oxidase subunit 1 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
534 a.a.
522 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00410
(COX2_YEAST) -
Cytochrome c oxidase subunit 2 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
251 a.a.
235 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00420
(COX3_YEAST) -
Cytochrome c oxidase subunit 3 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
269 a.a.
262 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P04037
(COX4_YEAST) -
Cytochrome c oxidase subunit 4, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
155 a.a.
49 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00424
(COX5A_YEAST) -
Cytochrome c oxidase subunit 5A, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
153 a.a.
96 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00427
(COX6_YEAST) -
Cytochrome c oxidase subunit 6, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
148 a.a.
99 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P10174
(COX7_YEAST) -
Cytochrome c oxidase subunit 7, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
60 a.a.
29 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P04039
(COX8_YEAST) -
Cytochrome c oxidase subunit 8, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
78 a.a.
31 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P07255
(COX9_YEAST) -
Cytochrome c oxidase subunit 9, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
59 a.a.
49 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
Q01519
(COX12_YEAST) -
Cytochrome c oxidase subunit 12, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
83 a.a.
57 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains C, D, E, N, O, P:
E.C.7.1.1.8
- quinol--cytochrome-c reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a quinol + 2 Fe(III)-[cytochrome c](out) = a quinone + 2 Fe(II)- [cytochrome c](out) + 2 H(+)(out)
|
 |
 |
 |
 |
 |
quinol
|
+
|
2
×
Fe(III)-[cytochrome c](out)
|
=
|
 quinone
quinone
|
+
|
2
×
Fe(II)- [cytochrome c](out)
|
+
|
2
×
H(+)(out)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains a, b, c:
E.C.7.1.1.9
- cytochrome-c oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4 Fe(II)-[cytochrome c] + O2 + 8 H+(in) = 4 Fe(III)-[cytochrome c] + 2 H2O + 4 H+(out)
|
 |
 |
 |
 |
 |
4
×
Fe(II)-[cytochrome c]
|
+
|
 2
×
O2
2
×
O2
|
+
|
8
×
H(+)(in)
|
=
|
4
×
Fe(III)-[cytochrome c]
|
+
|
2
×
H2O
|
+
|
4
×
H(+)(out)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chains d, e, f, g, h, i, j, k:
E.C.1.9.3.1
- Transferred entry: 7.1.1.9.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4 ferrocytochrome c + O2 + 4 H+ = 4 ferricytochrome c + 2 H2O
|
 |
 |
 |
 |
 |
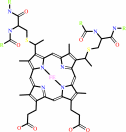 4
×
ferrocytochrome c
4
×
ferrocytochrome c
|
+
|
 2
×
O(2)
2
×
O(2)
|
+
|
4
×
H(+)
|
=
|
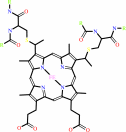 4
×
ferricytochrome c
4
×
ferricytochrome c
|
+
|
2
×
H(2)O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Mol Biol
26:50-57
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
Cryo-EM structure of the yeast respiratory supercomplex.
|
|
S.Rathore,
J.Berndtsson,
L.Marin-Buera,
J.Conrad,
M.Carroni,
P.Brzezinski,
M.Ott.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Respiratory chain complexes execute energy conversion by connecting electron
transport with proton translocation over the inner mitochondrial membrane to
fuel ATP synthesis. Notably, these complexes form multi-enzyme assemblies known
as respiratory supercomplexes. Here we used single-particle cryo-EM to determine
the structures of the yeast mitochondrial respiratory supercomplexes
III2IV and III2IV2, at 3.2-Å and 3.5-Å
resolutions, respectively. We revealed the overall architecture of the
supercomplex, which deviates from the previously determined assemblies in
mammals; obtained a near-atomic structure of the yeast complex IV; and
identified the protein-protein and protein-lipid interactions implicated in
supercomplex formation. Take together, our results demonstrate convergent
evolution of supercomplexes in mitochondria that, while building similar
assemblies, results in substantially different arrangements and structural
solutions to support energy conversion.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |

| | |

