 |
PDBsum entry 5int
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of thE C-terminal domain of coenzyme a biosynthesis bifunctional protein coabc
|
|
Structure:
|
 |
Phosphopantothenate--cysteine ligase. Chain: a, b. Synonym: phosphopantothenoylcysteine decarboxylase/phosphopantothenate--cysteine ligase,probable coenzyme a biosynthesis bifunctional protein coabc. Engineered: yes
|
|
Source:
|
 |
Bacillus anthracis. Organism_taxid: 1392. Gene: coabc, gbaa_4007, ab163_23415, ab164_18935, ab165_13850, ab166_05815, ab167_23710, ab168_27015, ab169_03120, ab170_11765, ab171_07935, ab893_19460, adk17_20550, adt20_07585, adt21_16720, bash2_01930. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.15Å
|
R-factor:
|
0.227
|
R-free:
|
0.258
|
|
|
Authors:
|
 |
B.Nocek,M.Zhou,S.Grimshaw,Y.Kim,W.F.Anderson,A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
B.Nocek
et al.
Crystal structure of the c-Terminal domain of coenzym biosynthesis bifunctional protein coabc.
To be published,
.
|
 |
|
Date:
|
 |
|
07-Mar-16
|
Release date:
|
06-Apr-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.4.1.1.36
- phosphopantothenoylcysteine decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Coenzyme A Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-[(R)-4-phosphopantothenoyl]-L-cysteine + H+ = (R)- 4'-phosphopantetheine + CO2
|
 |
 |
 |
 |
 |
N-[(R)-4-phosphopantothenoyl]-L-cysteine
|
+
|
H(+)
|
=
|
(R)- 4'-phosphopantetheine
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
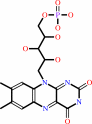 FMN
FMN
|
|
 |
 |
Enzyme class 2:
|
 |
Chains A, B:
E.C.6.3.2.5
- phosphopantothenate--cysteine ligase (CTP).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-4'-phosphopantothenate + L-cysteine + CTP = N-[(R)-4- phosphopantothenoyl]-L-cysteine + CMP + diphosphate + H+
|
 |
 |
 |
 |
 |
 (R)-4'-phosphopantothenate
(R)-4'-phosphopantothenate
|
+
|
 L-cysteine
L-cysteine
|
+
|
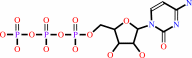 CTP
CTP
|
=
|
N-[(R)-4- phosphopantothenoyl]-L-cysteine
|
+
|
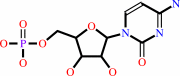 CMP
CMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

