 |
PDBsum entry 5foe
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of thE C. Elegans protein o-fucosyltransferase 2 (cepofut2) double mutant (r298k-r299k) in complex with gdp and the human tsr1 from thrombospondin 1
|
|
Structure:
|
 |
Gdp-fucose protein o-fucosyltransferase 2,thrombospondin-1. Chain: a, b. Synonym: patterning defective protein 2,peptide-o-fucosyltransferase 2,o-fuct-2. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Caenorhabditis elegans, homo sapiens. Human. Organism_taxid: 6239, 9606. Gene: pad-2, k10g9.3, thbs1, tsp, tsp1. Expressed in: komagataella pastoris. Expression_system_taxid: 4922.
|
|
Resolution:
|
 |
|
1.98Å
|
R-factor:
|
0.202
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
J.Valero-Gonzalez,C.Leonhard-Melief,E.Lira-Navarrete,G.Jimenez-Oses, C.Hernandez-Ruiz,M.C.Pallares,I.Yruela,D.Vasudevan,A.Lostao, F.Corzana,H.Takeuchi,R.S.Haltiwanger,R.Hurtado-Guerrero
|
|
Key ref:
|
 |
J.Valero-González
et al.
(2016).
A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2.
Nat Chem Biol,
12,
240-246.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
19-Nov-15
|
Release date:
|
27-Jan-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.1.221
- peptide-O-fucosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-seryl-[protein] + GDP-beta-L-fucose = 3-O-(alpha-L-fucosyl)-L- seryl-[protein] + GDP + H+
|
|
2.
|
L-threonyl-[protein] + GDP-beta-L-fucose = 3-O-(alpha-L-fucosyl)-L- threonyl-[protein] + GDP + H+
|
|
 |
 |
 |
 |
 |
L-seryl-[protein]
|
+
|
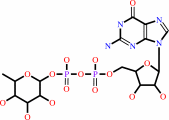 GDP-beta-L-fucose
GDP-beta-L-fucose
|
=
|
3-O-(alpha-L-fucosyl)-L- seryl-[protein]
|
+
|
GDP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
L-threonyl-[protein]
|
+
|
 GDP-beta-L-fucose
GDP-beta-L-fucose
|
=
|
3-O-(alpha-L-fucosyl)-L- threonyl-[protein]
|
+
|
GDP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Chem Biol
12:240-246
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2.
|
|
J.Valero-González,
C.Leonhard-Melief,
E.Lira-Navarrete,
G.Jiménez-Osés,
C.Hernández-Ruiz,
M.C.Pallarés,
I.Yruela,
D.Vasudevan,
A.Lostao,
F.Corzana,
H.Takeuchi,
R.S.Haltiwanger,
R.Hurtado-Guerrero.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Protein O-fucosyltransferase 2 (POFUT2) is an essential enzyme that fucosylates
serine and threonine residues of folded thrombospondin type 1 repeats (TSRs). To
date, the mechanism by which this enzyme recognizes very dissimilar TSRs has
been unclear. By engineering a fusion protein, we report the crystal structure
of Caenorhabditis elegans POFUT2 (CePOFUT2) in complex with GDP and human TSR1
that suggests an inverting mechanism for fucose transfer assisted by a catalytic
base and shows that nearly half of the TSR1 is embraced by CePOFUT2. A small
number of direct interactions and a large network of water molecules maintain
the complex. Site-directed mutagenesis demonstrates that POFUT2 fucosylates
threonine preferentially over serine and relies on folded TSRs containing the
minimal consensus sequence C-X-X-S/T-C. Crystallographic and mutagenesis data,
together with atomic-level simulations, uncover a binding mechanism by which
POFUT2 promiscuously recognizes the structural fingerprint of poorly homologous
TSRs through a dynamic network of water-mediated interactions.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

