 |
PDBsum entry 4npv

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/hydrolase inhibitor
|
PDB id
|
|

|

|
4npv
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase/hydrolase inhibitor
|
 |
|
Title:
|
 |
Crystal structure of human pde1b bound to inhibitor 7a (6,7,8- trimethoxy-n-(pentan-3-yl)quinazolin-4-amine)
|
|
Structure:
|
 |
Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1b. Chain: a. Fragment: unp residues 142-507. Synonym: cam-pde 1b, 63 kda cam-pde. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: pde1b, pde1b1, pdes1b. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.207
|
R-free:
|
0.236
|
|
|
Authors:
|
 |
J.Pandit,A.Evdomikov,M.Mansour,S.Simons
|
|
Key ref:
|
 |
J.M.Humphrey
et al.
Small-Molecule phosphodiesterase probes: discovery of and selective cns-Penetrable quinazoline inhibitors o.
Medchemcomm,
.
|
 |
|
Date:
|
 |
|
22-Nov-13
|
Release date:
|
16-Jul-14
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q01064
(PDE1B_HUMAN) -
Dual specificity calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
536 a.a.
323 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.17
- 3',5'-cyclic-nucleotide phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate + H+
|
 |
 |
 |
 |
 |
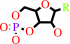 nucleoside 3',5'-cyclic phosphate
nucleoside 3',5'-cyclic phosphate
|
+
|
H2O
|
=
|
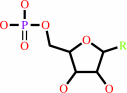 nucleoside 5'-phosphate
nucleoside 5'-phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

