
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 160 a.a.
160 a.a.
|
 |

|

|

|

|

|

|

|
 150 a.a.
150 a.a.
|
 |

|

|

|

|

|

|

|
 148 a.a.
148 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Viral protein/RNA
|
 |
|
Title:
|
 |
Crystal structure of macro domain of chikungunya virus in complex with RNA
|
|
Structure:
|
 |
Non-structural protein 3. Chain: a, b, c, d. Fragment: sequence database residues 1334-1493. Synonym: nsp3. Engineered: yes. RNA (5'-r( Ap Ap A)-3'). Chain: e, f. Engineered: yes
|
|
Source:
|
 |
Chikungunya virus. Organism_taxid: 371094. Strain: ross. Gene: nsp3. Expressed in: escherichia coli. Expression_system_taxid: 562. Synthetic: yes
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.218
|
R-free:
|
0.260
|
|
|
Authors:
|
 |
H.Malet,S.Jamal,B.Coutard,B.Canard
|
|
Key ref:
|
 |
H.Malet
et al.
(2009).
The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket.
J Virol,
83,
6534-6545.
PubMed id:
|
 |
|
Date:
|
 |
|
23-Mar-09
|
Release date:
|
21-Jul-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8JUX6
(POLN_CHIKS) -
Polyprotein P1234 from Chikungunya virus (strain S27-African prototype)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
2474 a.a.
160 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B, C, D:
E.C.2.1.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D:
E.C.2.7.7.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B, C, D:
E.C.2.7.7.19
- polynucleotide adenylyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
RNA(n) + ATP = RNA(n)-3'-adenine ribonucleotide + diphosphate
|
 |
 |
 |
 |
 |
 RNA(n)
RNA(n)
|
+
|
 ATP
ATP
|
=
|
RNA(n)-3'-adenine ribonucleotide
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chains A, B, C, D:
E.C.2.7.7.48
- RNA-directed Rna polymerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
RNA(n) + a ribonucleoside 5'-triphosphate = RNA(n+1) + diphosphate
|
 |
 |
 |
 |
 |
 RNA(n)
RNA(n)
|
+
|
ribonucleoside 5'-triphosphate
|
=
|
RNA(n+1)
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
Chains A, B, C, D:
E.C.3.1.3.84
- ADP-ribose 1''-phosphate phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
ADP-alpha-D-ribose 1''-phosphate + H2O = ADP-D-ribose + phosphate
|
 |
 |
 |
 |
 |
ADP-alpha-D-ribose 1''-phosphate
|
+
|
H2O
|
=
|
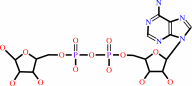 ADP-D-ribose
ADP-D-ribose
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
Chains A, B, C, D:
E.C.3.4.22.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 7:
|
 |
Chains A, B, C, D:
E.C.3.6.1.15
- nucleoside-triphosphate phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a ribonucleoside 5'-triphosphate + H2O = a ribonucleoside 5'-diphosphate + phosphate + H+
|
 |
 |
 |
 |
 |
ribonucleoside 5'-triphosphate
|
+
|
H2O
|
=
|
ribonucleoside 5'-diphosphate
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 8:
|
 |
Chains A, B, C, D:
E.C.3.6.1.74
- mRNA 5'-phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 5'-end triphospho-ribonucleoside in mRNA + H2O = a 5'-end diphospho- ribonucleoside in mRNA + phosphate + H+
|
 |
 |
 |
 |
 |
5'-end triphospho-ribonucleoside in mRNA
|
+
|
H2O
|
=
|
5'-end diphospho- ribonucleoside in mRNA
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 9:
|
 |
Chains A, B, C, D:
E.C.3.6.4.13
- Rna helicase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
ATP + H2O = ADP + phosphate + H+
|
 |
 |
 |
 |
 |
 ATP
ATP
|
+
|
H2O
|
=
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Virol
83:6534-6545
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket.
|
|
H.Malet,
B.Coutard,
S.Jamal,
H.Dutartre,
N.Papageorgiou,
M.Neuvonen,
T.Ahola,
N.Forrester,
E.A.Gould,
D.Lafitte,
F.Ferron,
J.Lescar,
A.E.Gorbalenya,
X.de Lamballerie,
B.Canard.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Macro domains (also called "X domains") constitute a protein module family
present in all kingdoms of life, including viruses of the Coronaviridae and
Togaviridae families. Crystal structures of the macro domain from the
Chikungunya virus (an "Old World" alphavirus) and the Venezuelan equine
encephalitis virus (a "New World" alphavirus) were determined at resolutions of
1.65 and 2.30 A, respectively. These domains are active as adenosine
di-phosphoribose 1''-phosphate phosphatases. Both the Chikungunya and the
Venezuelan equine encephalitis virus macro domains are ADP-ribose binding
modules, as revealed by structural and functional analysis. A single aspartic
acid conserved through all macro domains is responsible for the specific binding
of the adenine base. Sequence-unspecific binding to long, negatively charged
polymers such as poly(ADP-ribose), DNA, and RNA is observed and attributed to
positively charged patches outside of the active site pocket, as judged by
mutagenesis and binding studies. The crystal structure of the Chikungunya virus
macro domain with an RNA trimer shows a binding mode utilizing the same
adenine-binding pocket as ADP-ribose, but avoiding the ADP-ribose 1''-phosphate
phosphatase active site. This leaves the AMP binding site as the sole common
feature in all macro domains.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.F.Beitzel,
R.R.Bakken,
J.M.Smith,
and
C.S.Schmaljohn
(2010).
High-resolution functional mapping of the venezuelan equine encephalitis virus genome by insertional mutagenesis and massively parallel sequencing.
|
| |
PLoS Pathog,
6,
e1001146.
|
 |
|
|
|
|
 |
B.Morin,
B.Coutard,
M.Lelke,
F.Ferron,
R.Kerber,
S.Jamal,
A.Frangeul,
C.Baronti,
R.Charrel,
X.de Lamballerie,
C.Vonrhein,
J.Lescar,
G.Bricogne,
S.Günther,
and
B.Canard
(2010).
The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription.
|
| |
PLoS Pathog,
6,
0.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.C.Jaffar-Bandjee,
D.Ramful,
B.A.Gauzere,
J.J.Hoarau,
P.Krejbich-Trotot,
S.Robin,
A.Ribera,
J.Selambarom,
and
P.Gasque
(2010).
Emergence and clinical insights into the pathology of Chikungunya virus infection.
|
| |
Expert Rev Anti Infect Ther,
8,
987-996.
|
 |
|
|
|
|
 |
M.Karlsen,
S.Villoing,
K.F.Ottem,
E.Rimstad,
and
A.Nylund
(2010).
Development of infectious cDNA clones of Salmonid alphavirus subtype 3.
|
| |
BMC Res Notes,
3,
241.
|
 |
|
|
|
|
 |
M.Varjak,
E.Zusinaite,
and
A.Merits
(2010).
Novel functions of the alphavirus nonstructural protein nsP3 C-terminal region.
|
| |
J Virol,
84,
2352-2364.
|
 |
|
|
|
|
 |
E.Reichert,
A.Clase,
A.Bacetty,
and
J.Larsen
(2009).
Alphavirus antiviral drug development: scientific gap analysis and prospective research areas.
|
| |
Biosecur Bioterror,
7,
413-427.
|
 |
|
|
|
|
 |
J.A.Wojdyla,
I.Manolaridis,
E.J.Snijder,
A.E.Gorbalenya,
B.Coutard,
Y.Piotrowski,
R.Hilgenfeld,
and
P.A.Tucker
(2009).
Structure of the X (ADRP) domain of nsp3 from feline coronavirus.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1292-1300.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Jose,
J.E.Snyder,
and
R.J.Kuhn
(2009).
A structural and functional perspective of alphavirus replication and assembly.
|
| |
Future Microbiol,
4,
837-856.
|
 |
|
|
|
|
 |
M.Solignat,
B.Gay,
S.Higgs,
L.Briant,
and
C.Devaux
(2009).
Replication cycle of chikungunya: a re-emerging arbovirus.
|
| |
Virology,
393,
183-197.
|
 |
|
|
|
|
 |
P.Serrano,
M.A.Johnson,
A.Chatterjee,
B.W.Neuman,
J.S.Joseph,
M.J.Buchmeier,
P.Kuhn,
and
K.Wüthrich
(2009).
Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3.
|
| |
J Virol,
83,
12998-13008.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
|

