 |
PDBsum entry 2zvd

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.3
- triacylglycerol lipase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a triacylglycerol + H2O = a diacylglycerol + a fatty acid + H+
|
 |
 |
 |
 |
 |
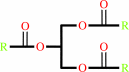 triacylglycerol
triacylglycerol
|
+
|
H2O
|
=
|
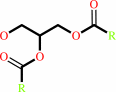 diacylglycerol
diacylglycerol
|
+
|
 fatty acid
fatty acid
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
400:82-95
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
X-ray crystallographic and MD simulation studies on the mechanism of interfacial activation of a family I.3 lipase with two lids.
|
|
C.Angkawidjaja,
H.Matsumura,
Y.Koga,
K.Takano,
S.Kanaya.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The interfacial activation mechanism of family I.3 lipase from Pseudomonas sp.
MIS38 (PML), which has two alpha-helical lids (lid1 and lid2), was investigated
using a combination of X-ray crystallography and molecular dynamics (MD)
simulation. The crystal structure of PML in an open conformation was determined
at 2.1 A resolution in the presence of Ca(2+) and Triton X-100. Comparison of
this structure with that in the closed conformation indicates that both lids
greatly change their positions and lid1 is anchored by the calcium ion (Ca1) in
the open conformation. This structure was not seriously changed even when the
protein was dialyzed extensively against the Ca(2+)-free buffer containing
Triton X-100 before crystallization, indicating that the open conformation is
fairly stable unless a micellar substance is removed. The crystal structure of
the PML derivative, in which the active site serine residue (Ser207) is
diethylphosphorylated by soaking the crystal of PML in the open conformation in
a solution containing diethyl p-nitrophenyl phosphate, was also determined. This
structure greatly resembles that in the open conformation, indicating that PML
structure in the open conformation represents that in the active form. MD
simulation of PML in the open conformation in the absence of micelles showed
that lid2 closes first, while lid1 maintains its open conformation. Likewise, MD
simulation of PML in the closed conformation in the absence of Ca(2+) and in the
presence of octane or trilaurin micelles showed that lid1 opens, while lid2
remains closed. These results suggest that Ca1 functions as a hook for
stabilization of a fully opened conformation of lid1 and for initiation of
subsequent opening of lid2.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Kuwahara,
C.Angkawidjaja,
Y.Koga,
K.Takano,
and
S.Kanaya
(2011).
Importance of an extreme C-terminal motif of a family I.3 lipase for stability.
|
| |
Protein Eng Des Sel,
24,
411-418.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

