 |
PDBsum entry 2ztg

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of archaeoglobus fulgidus alanyl-tRNA synthetase lacking thE C-terminal dimerization domain in complex with ala-sa
|
|
Structure:
|
 |
Alanyl-tRNA synthetase. Chain: a. Fragment: alars-deltac. Synonym: alanine-tRNA ligase, alars. Engineered: yes
|
|
Source:
|
 |
Archaeoglobus fulgidus. Organism_taxid: 2234. Gene: af_2255, alas. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.215
|
R-free:
|
0.264
|
|
|
Authors:
|
 |
M.Naganuma,S.Sekine,R.Fukunaga,S.Yokoyama,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
Key ref:

|
 |
M.Naganuma
et al.
(2009).
Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization.
Proc Natl Acad Sci U S A,
106,
8489-8494.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
01-Oct-08
|
Release date:
|
30-Jun-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O28029
(SYA_ARCFU) -
Alanine--tRNA ligase from Archaeoglobus fulgidus (strain ATCC 49558 / DSM 4304 / JCM 9628 / NBRC 100126 / VC-16)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
906 a.a.
739 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.7
- alanine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Ala) + L-alanine + ATP = L-alanyl-tRNA(Ala) + AMP + diphosphate
|
 |
 |
 |
 |
 |
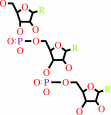 tRNA(Ala)
tRNA(Ala)
|
+
|
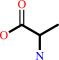 L-alanine
L-alanine
|
+
|
 ATP
ATP
|
=
|
L-alanyl-tRNA(Ala)
Bound ligand (Het Group name = )
matches with 59.38% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
106:8489-8494
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization.
|
|
M.Naganuma,
S.I.Sekine,
R.Fukunaga,
S.Yokoyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Alanyl-tRNA synthetase (AlaRS) specifically recognizes the major identity
determinant, the G3:U70 base pair, in the acceptor stem of tRNA(Ala) by both the
tRNA-recognition and editing domains. In this study, we solved the crystal
structures of 2 halves of Archaeoglobus fulgidus AlaRS: AlaRS-DeltaC, comprising
the aminoacylation, tRNA-recognition, and editing domains, and AlaRS-C,
comprising the dimerization domain. The aminoacylation/tRNA-recognition domains
contain an insertion incompatible with the class-specific tRNA-binding mode. The
editing domain is fixed tightly via hydrophobic interactions to the
aminoacylation/tRNA-recognition domains, on the side opposite from that in
threonyl-tRNA synthetase. A groove formed between the
aminoacylation/tRNA-recognition domains and the editing domain appears to be an
alternative tRNA-binding site, which might be used for the aminoacylation and/or
editing reactions. Actually, the amino acid residues required for the G3:U70
recognition are mapped in this groove. The dimerization domain consists of
helical and globular subdomains. The helical subdomain mediates dimerization by
forming a helix-loop-helix zipper. The globular subdomain, which is important
for the aminoacylation and editing activities, has a positively-charged face
suitable for tRNA binding.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
The aminoacylation and tRNA-recognition domains. (A) The
aminoacylation and tRNA-recognition domains of A. fulgidus
AlaRS, colored as in Fig. 1B, are shown. (B) The A. aeolicus
AlaRS-N structure, shown in the same orientation. The 2 regions
missing in A. fulgidus (InsB/E1 and InsB/E2) are colored brown,
and Mid2 is shown in gold.
|
 |
Figure 6.
A model of the full-length AlaRS dimer. Two copies of
AlaRS-ΔC, which are correlated by the crystallographic 2-fold
axis, and an AlaRS-C dimer, are shown. The N termini of AlaRS-C
were placed near the C termini of AlaRS-ΔC. The model was
colored as in Fig. 1.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Minajigi,
B.Deng,
and
C.S.Francklyn
(2011).
Fidelity escape by the unnatural amino acid β-hydroxynorvaline: an efficient substrate for Escherichia coli threonyl-tRNA synthetase with toxic effects on growth.
|
| |
Biochemistry,
50,
1101-1109.
|
 |
|
|
|
|
 |
M.Guo,
P.Schimmel,
and
X.L.Yang
(2010).
Functional expansion of human tRNA synthetases achieved by structural inventions.
|
| |
FEBS Lett,
584,
434-442.
|
 |
|
|
|
|
 |
M.Guo,
R.Shapiro,
P.Schimmel,
and
X.L.Yang
(2010).
Introduction of a leucine half-zipper engenders multiple high-quality crystals of a recalcitrant tRNA synthetase.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
243-250.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

