 |
PDBsum entry 2vz3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2vz3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Bleached galactose oxidase
|
|
Structure:
|
 |
Galactose oxidase. Chain: a. Synonym: gao, gaoase, go, bleached galactose oxidase. Engineered: yes
|
|
Source:
|
 |
Gibberella zeae. Organism_taxid: 5518. Expressed in: pichia pastoris. Expression_system_taxid: 4922.
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.169
|
R-free:
|
0.203
|
|
|
Authors:
|
 |
M.S.Rogers,R.Hurtado-Guerrero,S.J.Firbank,M.A.Halcrow,D.M.Dooley, S.E.V.Phillips,P.F.Knowles,M.J.Mcpherson
|
|
Key ref:
|
 |
M.S.Rogers
et al.
(2008).
Cross-link formation of the cysteine 228-tyrosine 272 catalytic cofactor of galactose oxidase does not require dioxygen.
Biochemistry,
47,
10428-10439.
PubMed id:
|
 |
|
Date:
|
 |
|
29-Jul-08
|
Release date:
|
16-Sep-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0CS93
(GAOA_GIBZA) -
Galactose oxidase from Gibberella zeae
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
680 a.a.
639 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.3.9
- galactose oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-galactose + O2 = D-galacto-hexodialdose + H2O2
|
 |
 |
 |
 |
 |
 D-galactose
D-galactose
|
+
|
 O2
O2
|
=
|
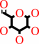 D-galacto-hexodialdose
D-galacto-hexodialdose
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
47:10428-10439
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Cross-link formation of the cysteine 228-tyrosine 272 catalytic cofactor of galactose oxidase does not require dioxygen.
|
|
M.S.Rogers,
R.Hurtado-Guerrero,
S.J.Firbank,
M.A.Halcrow,
D.M.Dooley,
S.E.Phillips,
P.F.Knowles,
M.J.McPherson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Galactose oxidase (GO) belongs to a class of proteins that self-catalyze
assembly of their redox-active cofactors from active site amino acids.
Generation of enzymatically active GO appears to require at least four
sequential post-translational modifications: cleavage of a secretion signal
sequence, copper-dependent cleavage of an N-terminal pro sequence,
copper-dependent formation of a C228-Y272 thioether bond, and generation of the
Y272 radical. The last two processes were investigated using a truncated protein
(termed premat-GO) lacking the pro sequence and purified under copper-free
conditions. Reactions of premat-GO with Cu(II) were investigated using optical,
EPR, and resonance Raman spectroscopy, SDS-PAGE, and X-ray crystallography.
Premat-GO reacted anaerobically with excess Cu(II) to efficiently form the
thioether bond but not the Y272 radical. A potential C228-copper coordinated
intermediate (lambda max = 406 nm) in the processing reaction, which had not yet
formed the C228-Y272 cross-link, was identified from the absorption spectrum. A
copper-thiolate protein complex, with copper coordinated to C228, H496, and
H581, was also observed in a 3 min anaerobic soak by X-ray crystallography,
whereas a 24 h soak revealed the C228-Y272 thioether bond. In solution, addition
of oxygenated buffer to premat-GO preincubated with excess Cu(II) generated the
Y272 radical state. On the basis of these data, a mechanism for the formation of
the C228-Y272 bond and tyrosyl radical generation is proposed. The 406 nm
complex is demonstrated to be a catalytically competent processing intermediate
under anaerobic conditions. We propose a potential mechanism which is in common
with aerobic processing by Cu(II) until the step at which the second electron
acceptor is required.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.H.Stipanuk,
C.R.Simmons,
P.Andrew Karplus,
and
J.E.Dominy
(2011).
Thiol dioxygenases: unique families of cupin proteins.
|
| |
Amino Acids,
41,
91.
|
 |
|
|
|
|
 |
O.Spadiut,
L.Olsson,
and
H.Brumer
(2010).
A comparative summary of expression systems for the recombinant production of galactose oxidase.
|
| |
Microb Cell Fact,
9,
68.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

