 |
PDBsum entry 2jb4

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2jb4
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.21.3.1
- isopenicillin-N synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Penicillin N and Deacetoxycephalosporin C Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine + O2 = isopenicillin N + 2 H2O
|
 |
 |
 |
 |
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine
|
+
|
 O2
O2
|
=
|
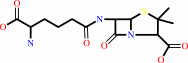 isopenicillin N
isopenicillin N
|
+
|
2
×
H2O
Bound ligand (Het Group name = )
matches with 92.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Chembiochem
8:2003-2007
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
A cyclobutanone analogue mimics penicillin in binding to isopenicillin N synthase.
|
|
A.C.Stewart,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
P.J.Rutledge.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A carbocyclic analogue of the beta-lactam antibiotic isopenicillin N (IPN) has
been synthesised and cocrystallised with isopenicillin N synthase (IPNS), the
central enzyme in the biosynthesis of penicillin antibiotics. The crystal
structure of the IPNS-cyclobutanone complex reveals an active site environment
similar to that seen in the enzyme-product complex generated by turnover of the
natural substrate within the crystalline protein. The IPNS-cyclobutanone
structure demonstrates that the product analogue is tethered to the protein by
hydrogen bonding and salt bridge interactions with its carboxylate groups, as
seen previously for the natural substrate and product. Furthermore, the
successful cocrystallisation of this analogue with IPNS provides firm structural
evidence for the utility of such cyclobutanone derivatives as hydrolytically
stable analogues of bicyclic beta-lactams.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
W.Ge,
I.J.Clifton,
A.R.Howard-Jones,
J.E.Stok,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2009).
Structural studies on the reaction of isopenicillin N synthase with a sterically demanding depsipeptide substrate analogue.
|
| |
Chembiochem,
10,
2025-2031.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

