 |
PDBsum entry 2eob

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Solution structure of the second sh2 domain from rat plc gamma-2
|
|
Structure:
|
 |
1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 2. Chain: a. Fragment: sh2 domain. Synonym: phosphoinositide phospholipasE C, plc-gamma-2, phospholipasE C-gamma-2, plc-iv. Engineered: yes
|
|
Source:
|
 |
Rattus norvegicus. Norway rat. Organism_taxid: 10116. Gene: plcg2. Other_details: cell-free protein synthesis
|
|
NMR struc:
|
 |
20 models
|
 |
|
Authors:
|
 |
R.Sano,F.Hayashi,T.Nagashima,M.Yoshida,S.Yokoyama,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
|
Key ref:
|
 |
R.Sano
et al.
Solution structure of the second sh2 domain from rat plc gamma-2.
To be published,
.
|
 |
|
Date:
|
 |
|
29-Mar-07
|
Release date:
|
01-Apr-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P24135
(PLCG2_RAT) -
1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2 from Rattus norvegicus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1265 a.a.
124 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 13 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.11
- phosphoinositide phospholipase C.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
myo-Inositol Phosphate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate) + H2O = 1D-myo-inositol 1,4,5-trisphosphate + a 1,2-diacyl-sn-glycerol + H+
|
 |
 |
 |
 |
 |
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate)
|
+
|
H2O
|
=
|
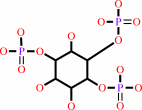 1D-myo-inositol 1,4,5-trisphosphate
1D-myo-inositol 1,4,5-trisphosphate
|
+
|
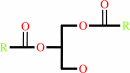 1,2-diacyl-sn-glycerol
1,2-diacyl-sn-glycerol
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

