 |
PDBsum entry 2ca6

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Signaling regulator
|
PDB id
|
|

|

|
2ca6
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Detecting and overcoming hemihedral twinning during the mir structure determination of RNA1p.
|
 |
|
Authors
|
 |
R.C.Hillig,
L.Renault.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2006,
62,
750-765.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
The structure of Rna1p was originally solved to 2.7 A resolution by MIRAS from
crystals with partial hemihedral twinning in space group I4(1) [Hillig et al.
(1999), Mol. Cell, 3, 781-791] by finding a low-twinned native crystal (twin
fraction alpha=0.06) and after twin correction of all data sets. Rna1p crystals
have now been used to examine how far twinning and twin correction affect MIR
phasing with a higher resolution but highly twinned native data set. Even high
hemihedral twinning [alphanative=0.39, alphaderivative=0.24] would not have
hindered heavy-atom site identification of strong derivatives using difference
Patterson maps. However, a weaker derivative could have been missed and
refinement would have stalled at high R values had twinning not been identified
and accounted for. Twin correction improved both site identification,
experimental phasing statistics and MIR map quality. Different strategies were
tested for refinement against twinned data. Using uncorrected twinned data and
TWIN-CNS, Rna1p has now been refined to 2.2 A resolution (final twinned R and
Rfree were 0.165 and 0.218, respectively). The increased resolution enabled
release of the NCS restraints and allowed new conclusions to be drawn on the
flexibility of the two molecules in the asymmetric unit. In the case of Rna1p,
twinned crystal growth was possible owing to the presence of a twofold NCS axis
almost parallel to the twin operator.
|
 |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Crystals and diffraction pattern of S. pombe Rna1p.
(a) Typical sea-urchin-like crystal clusters. (b) Manually
separated crystals of about 600 × 40 × 40 µm.
(c) Diffraction pattern from native-A (high-resolution sweep,
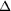 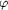 =
0.5°, exposure time 8 min, detector edge 2 Å). Yellow
boxes denote the enlarged regions. The reflections show no signs
of splitting. =
0.5°, exposure time 8 min, detector edge 2 Å). Yellow
boxes denote the enlarged regions. The reflections show no signs
of splitting.
|
 |
Figure 6.
Figure 6 Structure of Rna1p refined to 2.20 Å. (a)
Representative view of the final 3F[o] - 2F[c] electron-density
map. Shown is the region around leucine-rich repeat 8 (LRR8),
contoured at 1.5  .
(b) Ribbon representation of Rna1p. (c) Superimposition of the
two independent molecules A (red) and B (blue) in the asymmetric
unit (C^ .
(b) Ribbon representation of Rna1p. (c) Superimposition of the
two independent molecules A (red) and B (blue) in the asymmetric
unit (C^  backbone
representation). Differences are found in the N-terminal region
as well as in LRR3/LRR4. (d) Superimposition of Rna1p (molecules
A and B in red and blue, respectively) and the complex of Rna1p
(green) with Ran-GMPPNP-Mg-RanBP1 (PDB code 1k5d ). The Ran
backbone is shown as a grey ribbon, GMPPNP in stick
representation and Mg as magenta-coloured sphere. An enlargement
of the region of the flexible loops of LRR3/LRR4 is shown. This
region, which differs between molecules A and B in the
high-resolution structure of Rna1p, coincides with part of the
interface between Rna1p and Ran-GMPPNP. The flexibility in Rna1p
may indicate an inherent mobility designed to allow an induced
fit. backbone
representation). Differences are found in the N-terminal region
as well as in LRR3/LRR4. (d) Superimposition of Rna1p (molecules
A and B in red and blue, respectively) and the complex of Rna1p
(green) with Ran-GMPPNP-Mg-RanBP1 (PDB code 1k5d ). The Ran
backbone is shown as a grey ribbon, GMPPNP in stick
representation and Mg as magenta-coloured sphere. An enlargement
of the region of the flexible loops of LRR3/LRR4 is shown. This
region, which differs between molecules A and B in the
high-resolution structure of Rna1p, coincides with part of the
interface between Rna1p and Ran-GMPPNP. The flexibility in Rna1p
may indicate an inherent mobility designed to allow an induced
fit.
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2006,
62,
750-765)
copyright 2006.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
The crystal structure of RNA1p: a new fold for a gtpase-Activating protein.
|
 |
|
Authors
|
 |
R.C.Hillig,
L.Renault,
I.R.Vetter,
T.Drell,
A.Wittinghofer,
J.Becker.
|
 |
|
Ref.
|
 |
Mol Cell, 1999,
3,
781-791.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. NCS-Averaged Experimental Electron Density MapPart
of the map showing the region of LRR8 (residues 217–243). The
refined model (ball-and-stick) is superimposed on the
experimental electron density map after 2-fold NCS averaging,
solvent flattening, and histogram matching, contoured at 1.0 σ.
Figure prepared using Bobscript ([28]).
|
 |
Figure 4.
Figure 4. Structural Overlay of the 11 LRRs of rna1pLRRs that
deviate markedly from the ideal LRR structure are presented in
blue (LRR1), red (LRR3), and green (LRR5). The position of
Arg-74 is marked by a red ball. The LRR consensus residues of
LRR8 are depicted as ball-and-stick models (yellow) illustrating
the formation of the hydrophobic core. Figure produced with
Bobscript ([28]).
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from Cell Press
|
 |
|
Secondary reference #2
|
 |
|
Title
|
 |
Rangap mediates gtp hydrolysis without an arginine finger.
|
 |
|
Authors
|
 |
M.J.Seewald,
C.Körner,
A.Wittinghofer,
I.R.Vetter.
|
 |
|
Ref.
|
 |
Nature, 2002,
415,
662-666.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2: Details of the Ran–RanGAP interface. a, Worm plot
of Ran and RanGAP, with colours as in Fig. 1 and important
residues shown in ball and stick representation. The uncomplexed
RanGAP (cyan) is superimposed; arrows indicate the movement of
Lys 76 and Arg 74, and asterisk symbolizes the potential clash
of Leu 43 and Lys 76 on complex formation. b, Switch II of Ran
(green) superimposed on the Ran–ranBD1 structure (blue),
illustrating the reorientation of Gln 69 into a catalytically
competent conformation by the residues Tyr 39 from Ran and Asn
131 fron RanGAP.
|
 |
Figure 3.
Figure 3: The active site. a, The nucleotide and relevant
residues of Ran and RanGAP; interactions are indicated by dashed
lines. The catalytic water ('W') has very weak density (probably
owing to the limited resolution) and was not included in the
final model, but is shown here to illustrate its potential
interaction partners. It sits in a similar position as in the
Ras–RasGAP complex. Asterisk denotes the hydroxyl group of Thr
42 of Ran. The rest of the side chain is omitted for clarity. b,
The 2F[o] - F[c] electron density map contoured at 1.6  for
the structure with GDP and aluminium fluoride, the latter
modelled as AlF[3]. for
the structure with GDP and aluminium fluoride, the latter
modelled as AlF[3].
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from Macmillan Publishers Ltd
|
 |
|

|
|
|
 |

