 |
PDBsum entry 2bhc
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Structural and functional implications of metal ion selection in aminopeptidase p, A metalloprotease with a dinuclear metal center.
|
 |
|
Authors
|
 |
S.C.Graham,
C.S.Bond,
H.C.Freeman,
J.M.Guss.
|
 |
|
Ref.
|
 |
Biochemistry, 2005,
44,
13820-13836.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
The effect of metal substitution on the activity and structure of the
aminopeptidase P (APPro) from Escherichia coli has been investigated.
Measurements of activity in the presence of Mn2+, Mg2+, Zn2+, Na+, and Ca2+ show
that significant activity is seen only in the Mn-bound form of the enzyme. The
addition of Zn2+ to [MnMn(APPro)] is strongly inhibitory. Crystal structures of
[MnMn(APPro)], [MgMg(APPro)], [ZnZn(APPro)], [ZnMg(APPro)], [Ca_(APPro)],
[Na_(APPro)], and [apo(APPro)] were determined. The structures of [Ca_(APPro)]
and [Na_(APPro)] have a single metal atom at their active site. Surprisingly,
when a tripeptide substrate (ValProLeu) was soaked into [Na_(APPro)] crystals in
the presence of 200 mM Mg2+, the structure had substrate, but no metal, bound at
the active site. The structure of apo APPro complexed with ValProLeu shows that
the N-terminal amino group of a substrate can be bound at the active site by
carboxylate side chains that normally bind the second metal atom, providing a
model for substrate binding in a single-metal active enzyme. Structures of
[MnMn(APPro)] and [ZnZn(APPro)] complexes of ProLeu, a product inhibitor, in the
presence of excess Zn reveal a third metal-binding site, formed by two conserved
His residues and the dipeptide inhibitor. A Zn atom bound at such a site would
stabilize product binding and enhance inhibition.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
An orthorhombic form of escherichia coli aminopeptidase p at 2.4 a resolution.
|
 |
|
Authors
|
 |
S.C.Graham,
M.Lee,
H.C.Freeman,
J.M.Guss.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2003,
59,
897-902.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Projection of the six independent subunits in the
asymmetric unit of orthorhombic AMPP. The red, green, blue and
yellow subunits form a tetramer with approximate
(non-crystallographic) 222 symmetry. The remaining subunits are
half of a second tetramer formed by the operation of a
crystallographic twofold parallel to the vertical edge of the
paper. The colour in the one of these subunits has been ramped
from blue at the N-terminus to red at the C-terminus to
facilitate the distinction between the domains.
|
 |
Figure 2.
Figure 2 A preferred orientation of the imidazole ring of
His350, showing hydrogen bonds to the O(peptide) of His351 and
to the O 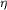 of
Tyr387. Two nearby ligands of Mn[A] and their interactions with
the metal are also shown. of
Tyr387. Two nearby ligands of Mn[A] and their interactions with
the metal are also shown.
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from the IUCr
|
 |
|

|
|
|
 |

