 |
PDBsum entry 2ad5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.2
- Ctp synthase (glutamine hydrolyzing).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
UTP + L-glutamine + ATP + H2O = CTP + L-glutamate + ADP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
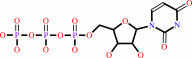 UTP
UTP
|
+
|
 L-glutamine
L-glutamine
|
+
|
 ATP
ATP
|
+
|
H2O
|
=
|
CTP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
L-glutamate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
44:13491-13499
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mechanisms of product feedback regulation and drug resistance in cytidine triphosphate synthetases from the structure of a CTP-inhibited complex.
|
|
J.A.Endrizzi,
H.Kim,
P.M.Anderson,
E.P.Baldwin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cytidine triphosphate synthetases (CTPSs) synthesize CTP and regulate its
intracellular concentration through direct interactions with the four
ribonucleotide triphosphates. In particular, CTP product is a feedback inhibitor
that competes with UTP substrate. Selected CTPS mutations that impart resistance
to pyrimidine antimetabolite inhibitors also relieve CTP inhibition and cause a
dramatic increase in intracellular CTP concentration, indicating that the drugs
act by binding to the CTP inhibitory site. Resistance mutations map to a pocket
that, although adjacent, does not coincide with the expected UTP binding site in
apo Escherichia coli CTPS [EcCTPS; Endrizzi, J. A., et al. (2004) Biochemistry
43, 6447-6463], suggesting allosteric rather than competitive inhibition. Here,
bound CTP and ADP were visualized in catalytically active EcCTPS crystals soaked
in either ATP and UTP substrates or ADP and CTP products. The CTP cytosine ring
resides in the pocket predicted by the resistance mutations, while the
triphosphate moiety overlaps the putative UTP triphosphate binding site,
explaining how CTP competes with UTP while CTP resistance mutations are acquired
without loss of catalytic efficiency. Extensive complementarity and interaction
networks at the interfacial binding sites provide the high specificity for
pyrimidine triphosphates and mediate nucleotide-dependent tetramer formation.
Overall, these results depict a novel product inhibition strategy in which
shared substrate and product moieties bind to a single subsite while specificity
is conferred by separate subsites. This arrangement allows for independent
adaptation of UTP and CTP binding affinities while efficiently utilizing the
enzyme surface.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.L.Liu
(2011).
The enigmatic cytoophidium: Compartmentation of CTP synthase via filament formation.
|
| |
Bioessays,
33,
159-164.
|
 |
|
|
|
|
 |
C.Noree,
B.K.Sato,
R.M.Broyer,
and
J.E.Wilhelm
(2010).
Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster.
|
| |
J Cell Biol,
190,
541-551.
|
 |
|
|
|
|
 |
F.A.Lunn,
J.E.Macdonnell,
and
S.L.Bearne
(2008).
Structural Requirements for the Activation of Escherichia coli CTP Synthase by the Allosteric Effector GTP Are Stringent, but Requirements for Inhibition Are Lax.
|
| |
J Biol Chem,
283,
2010-2020.
|
 |
|
|
|
|
 |
S.D.Taylor,
F.A.Lunn,
and
S.L.Bearne
(2008).
Ground state, intermediate, and multivalent nucleotide analogue inhibitors of cytidine 5'-triphosphate synthase.
|
| |
ChemMedChem,
3,
1853-1857.
|
 |
|
|
|
|
 |
Y.F.Chang,
and
G.M.Carman
(2008).
CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae.
|
| |
Prog Lipid Res,
47,
333-339.
|
 |
|
|
|
|
 |
Y.F.Chang,
S.S.Martin,
E.P.Baldwin,
and
G.M.Carman
(2007).
Phosphorylation of human CTP synthetase 1 by protein kinase C: identification of Ser(462) and Thr(455) as major sites of phosphorylation.
|
| |
J Biol Chem,
282,
17613-17622.
|
 |
|
|
|
|
 |
P.Kursula,
S.Flodin,
M.Ehn,
M.Hammarström,
H.Schüler,
P.Nordlund,
and
P.Stenmark
(2006).
Structure of the synthetase domain of human CTP synthetase, a target for anticancer therapy.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
613-617.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

