 |
PDBsum entry 2wjz

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase/transferase
|
PDB id
|
|

|

|
2wjz
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 251 a.a.
251 a.a.
|
 |

|

|

|

|

|

|

|


 193 a.a.
193 a.a.
|
 |

|

|

|

|

|

|

|
 237 a.a.
237 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase/transferase
|
 |
|
Title:
|
 |
Crystal structure of (hish) k181a y138a mutant of imidazoleglycerolphosphate synthase (hish hisf) which displays constitutive glutaminase activity
|
|
Structure:
|
 |
Imidazole glycerol phosphate synthase hisf. Chain: a, c, e. Synonym: igp synthase cyclase subunit, igp synthase subunit hisf, imgp synthase subunit hisf, igps subunit hisf. Engineered: yes. Imidazole glycerol phosphate synthase subunit hish. Chain: b, d, f. Synonym: igp synthase glutamine amidotransferase subunit, igp synthase subunit hish, imgp synthase subunit hish, tmhish, igps
|
|
Source:
|
 |
Thermotoga maritima. Organism_taxid: 2336. Expressed in: escherichia coli. Expression_system_taxid: 469008. Expressed in: escherichia coli k-12. Expression_system_taxid: 83333.
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.187
|
R-free:
|
0.218
|
|
|
Authors:
|
 |
M.C.Vega,F.List,A.Razeto,M.C.Haeger,K.Babinger,J.Kuper,R.Sterner, M.Wilmanns
|
|
Key ref:
|
 |
F.List
et al.
(2012).
Catalysis uncoupling in a glutamine amidotransferase bienzyme by unblocking the glutaminase active site.
Chem Biol,
19,
1589-1599.
PubMed id:
|
 |
|
Date:
|
 |
|
02-Jun-09
|
Release date:
|
25-Aug-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9X0C6
(HIS6_THEMA) -
Imidazole glycerol phosphate synthase subunit HisF from Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
253 a.a.
251 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D, E, F:
E.C.4.3.2.10
- imidazole glycerol-phosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-[(5-phospho-1-deoxy-D-ribulos-1-ylimino)methylamino]-1-(5-phospho-beta- D-ribosyl)imidazole-4-carboxamide + L-glutamine = D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate + 5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide + L-glutamate + H+
|
 |
 |
 |
 |
 |
5-[(5-phospho-1-deoxy-D-ribulos-1-ylimino)methylamino]-1-(5-phospho-beta- D-ribosyl)imidazole-4-carboxamide
|
+
|
 L-glutamine
L-glutamine
|
=
|
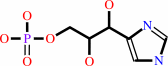 D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate
D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate
|
+
|
5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide
|
+
|
 L-glutamate
L-glutamate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains B, D, F:
E.C.3.5.1.2
- glutaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutamine + H2O = L-glutamate + NH4+
|
 |
 |
 |
 |
 |
 L-glutamine
L-glutamine
|
+
|
H2O
|
=
|
 L-glutamate
L-glutamate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Chem Biol
19:1589-1599
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Catalysis uncoupling in a glutamine amidotransferase bienzyme by unblocking the glutaminase active site.
|
|
F.List,
M.C.Vega,
A.Razeto,
M.C.Häger,
R.Sterner,
M.Wilmanns.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| |

