 |
PDBsum entry 2vzk

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|



 173 a.a.
173 a.a.
|
 |

|

|

|

|

|

|

|

 213 a.a.
213 a.a.
|
 |

|

|

|

|

|

|

|

 197 a.a.
197 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structure of the acyl-enzyme complex of an n-terminal nucleophile (ntn) hydrolase, oat2
|
|
Structure:
|
 |
Glutamate n-acetyltransferase 2 alpha chain. Chain: a, c, e, g. Fragment: residues 8-180. Synonym: ornithine acetyl transferase, oat2. Engineered: yes. Glutamate n-acetyltransferase 2 beta chain. Chain: b. Synonym: ornithine acetyl transferase, oat2. Engineered: yes.
|
|
Source:
|
 |
Streptomyces clavuligerus. Organism_taxid: 1901. Atcc: 3585. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.33Å
|
R-factor:
|
0.255
|
R-free:
|
0.284
|
|
|
Authors:
|
 |
A.Iqbal,I.J.Clifton,C.J.Schofield
|
|
Key ref:
|
 |
A.Iqbal
et al.
(2009).
Anatomy of a simple acyl intermediate in enzyme catalysis: combined biophysical and modeling studies on ornithine acetyl transferase.
J Am Chem Soc,
131,
749-757.
PubMed id:
|
 |
|
Date:
|
 |
|
01-Aug-08
|
Release date:
|
16-Sep-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0DJQ5
(GNAT2_STRCL) -
Glutamate N-acetyltransferase 2 from Streptomyces clavuligerus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
393 a.a.
173 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H:
E.C.2.3.1.35
- glutamate N-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N2-acetyl-L-ornithine + L-glutamate = N-acetyl-L-glutamate + L-ornithine
|
 |
 |
 |
 |
 |
N(2)-acetyl-L-ornithine
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
 L-glutamate
L-glutamate
|
=
|
N-acetyl-L-glutamate
Bound ligand (Het Group name = )
matches with 41.67% similarity
|
+
|
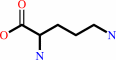 L-ornithine
L-ornithine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Am Chem Soc
131:749-757
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Anatomy of a simple acyl intermediate in enzyme catalysis: combined biophysical and modeling studies on ornithine acetyl transferase.
|
|
A.Iqbal,
I.J.Clifton,
M.Bagonis,
N.J.Kershaw,
C.Domene,
T.D.Claridge,
C.W.Wharton,
C.J.Schofield.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Acyl-enzyme complexes are intermediates in reactions catalyzed by many
hydrolases and related enzymes which employ nucleophilic catalysis. However,
most of the reported structural data on acyl-enzyme complexes has been acquired
under noncatalytic conditions. Recent IR analyses have indicated that some
acyl-enzyme complexes may be more flexible than most crystallographic analyses
have implied. OAT2 is a member of the N-terminal nucleophile (Ntn) hydrolase
enzyme superfamily and catalyzes the reversible transfer of an acetyl group
between the alpha-amino groups of ornithine and glutamate in a mechanism
proposed to involve an acyl-enzyme complex. We have carried out biophysical
analyses on ornithine acetyl transferase (OAT2), both in solution and in the
crystalline state. Mass spectrometric studies identified Thr-181 as the residue
acetylated during OAT2 catalysis; (13)C NMR analyses implied the presence of an
acyl-enzyme complex in solution. Crystallization of OAT2 in the presence of
N-alpha-acetyl-L-glutamate led to a structure in which Thr-181 was acetylated;
the carbonyl oxygen of the acyl-enzyme complex was located in an oxyanion hole
and positioned to hydrogen bond with the backbone amide NH of Gly-112 and the
alcohol of Thr-111. While the crystallographic analyses revealed only one
structure, IR spectroscopy demonstrated the presence of two distinct acyl-enzyme
complex structures with carbonyl stretching frequencies at 1691 and 1701 cm(-1).
Modeling studies implied two possible acyl-enzyme complex structures, one of
which correlates with that observed in the crystal structure and with the 1691
cm(-1) IR absorption. The second acyl-enzyme complex structure, which has only a
single oxyanion hole hydrogen bond, is proposed to give rise to the 1701 cm(-1)
IR absorption. The two acyl-enzyme complex structures can interconvert by
movement of the Thr-111 side-chain alcohol hydrogen away from the oxyanion hole
to hydrogen bond with the backbone carbonyl of the acylated residue, Thr-181.
Overall, the results reveal that acyl-enzyme complex structures may be more
dynamic than previously thought and support the use of a comprehensive
biophysical and modeling approach in studying such intermediates.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Hebecker,
W.Arendt,
I.U.Heinemann,
J.H.Tiefenau,
M.Nimtz,
M.Rohde,
D.Söll,
and
J.Moser
(2011).
Alanyl-phosphatidylglycerol synthase: mechanism of substrate recognition during tRNA-dependent lipid modification in Pseudomonas aeruginosa.
|
| |
Mol Microbiol,
80,
935-950.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
| |

