 |
PDBsum entry 2v4i

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structure of a novel n-acyl-enzyme intermediate of an n-terminal nucleophile (ntn) hydrolase, oat2
|
|
Structure:
|
 |
Glutamate n-acetyltransferase 2 alpha chain. Chain: a, c, e, g. Fragment: residues 8-180. Synonym: ornithine acetyl transferase, oat2. Engineered: yes. Glutamate n-acetyltransferase 2 beta chain. Chain: b, d, f, h. Synonym: ornithine acetyl transferase, oat2. Engineered: yes.
|
|
Source:
|
 |
Streptomyces clavuligerus. Organism_taxid: 1901. Atcc: 3585. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.216
|
R-free:
|
0.247
|
|
|
Authors:
|
 |
A.Iqbal,I.J.Clifton,C.J.Schofield
|
|
Key ref:
|
 |
A.Iqbal
et al.
Structure of a novel n-Acyl-Enzyme intermediate of an n-Terminal nucleophile (ntn) hydrolase, Oat2.
To be published,
.
|
 |
|
Date:
|
 |
|
22-Sep-08
|
Release date:
|
07-Oct-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H:
E.C.2.3.1.35
- glutamate N-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N2-acetyl-L-ornithine + L-glutamate = N-acetyl-L-glutamate + L-ornithine
|
 |
 |
 |
 |
 |
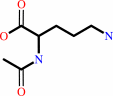 N(2)-acetyl-L-ornithine
N(2)-acetyl-L-ornithine
|
+
|
 L-glutamate
L-glutamate
|
=
|
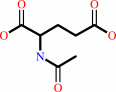 N-acetyl-L-glutamate
N-acetyl-L-glutamate
|
+
|
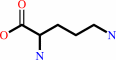 L-ornithine
L-ornithine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

