 |
PDBsum entry 1z8s

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.101
- Dna primase DnaG.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
ssDNA + n NTP = ssDNA/pppN(pN)n-1 hybrid + (n-1) diphosphate
|
 |
 |
 |
 |
 |
ssDNA
|
+
|
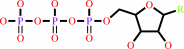 n
NTP
n
NTP
|
=
|
ssDNA/pppN(pN)n-1 hybrid
|
+
|
(n-1) diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
13:609-616
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Solution structure of the helicase-interaction domain of the primase DnaG: a model for helicase activation.
|
|
K.Syson,
J.Thirlway,
A.M.Hounslow,
P.Soultanas,
J.P.Waltho.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The helicase-primase interaction is a critical event in DNA replication and is
mediated by a putative helicase-interaction domain within the primase. The
solution structure of the helicase-interaction domain of DnaG reveals that it is
made up of two independent subdomains: an N-terminal six-helix module and a
C-terminal two-helix module that contains the residues of the primase previously
identified as important in the interaction with the helicase. We show that the
two-helix module alone is sufficient for strong binding between the primase and
the helicase but fails to activate the helicase; both subdomains are required
for helicase activation. The six-helix module of the primase has only one close
structural homolog, the N-terminal domain of the corresponding helicase. This
surprising structural relationship, coupled with the differences in surface
properties of the two molecules, suggests how the helicase-interaction domain
may perturb the structure of the helicase and lead to activation.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 6.
Figure 6. A Model for the DnaB-P16 Interaction
(A) The
3-fold symmetric ring of hexameric DnaB, showing the N-domain of
one monomer (6N) interacting with the C-domain (5H) of the
neighboring monomer (Yang et al., 2003).
(B) In the
DnaB-P16 complex, the P16 protein (shaded purple) interacts with
the linker region that connects the N-terminal (6N) and
C-terminal (6H) domains of one monomer, via its C2 subdomain. In
addition, the C1 subdomain displaces 6N while at the same time
maintaining the interactions with 5H that are essential to
preserve a 3-fold symmetric ring in the helicase-primase complex.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2005,
13,
609-616)
copyright 2005.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Li,
J.Liu,
L.Zhou,
H.Pei,
J.Zhou,
and
H.Xiang
(2010).
Two distantly homologous DnaG primases from Thermoanaerobacter tengcongensis exhibit distinct initiation specificities and priming activities.
|
| |
J Bacteriol,
192,
2670-2681.
|
 |
|
|
|
|
 |
M.A.Larson,
M.A.Griep,
R.Bressani,
K.Chintakayala,
P.Soultanas,
and
S.H.Hinrichs
(2010).
Class-specific restrictions define primase interactions with DNA template and replicative helicase.
|
| |
Nucleic Acids Res,
38,
7167-7178.
|
 |
|
|
|
|
 |
G.Wang,
M.G.Klein,
E.Tokonzaba,
Y.Zhang,
L.G.Holden,
and
X.S.Chen
(2008).
The structure of a DnaB-family replicative helicase and its interactions with primase.
|
| |
Nat Struct Mol Biol,
15,
94.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Chintakayala,
M.A.Larson,
M.A.Griep,
S.H.Hinrichs,
and
P.Soultanas
(2008).
Conserved residues of the C-terminal p16 domain of primase are involved in modulating the activity of the bacterial primosome.
|
| |
Mol Microbiol,
68,
360-371.
|
 |
|
|
|
|
 |
S.A.Koepsell,
M.A.Larson,
C.A.Frey,
S.H.Hinrichs,
and
M.A.Griep
(2008).
Staphylococcus aureus primase has higher initiation specificity, interacts with single-stranded DNA stronger, but is less stimulated by its helicase than Escherichia coli primase.
|
| |
Mol Microbiol,
68,
1570-1582.
|
 |
|
|
|
|
 |
T.Biswas,
and
O.V.Tsodikov
(2008).
Hexameric ring structure of the N-terminal domain of Mycobacterium tuberculosis DnaB helicase.
|
| |
FEBS J,
275,
3064-3071.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Chintakayala,
M.A.Larson,
W.H.Grainger,
D.J.Scott,
M.A.Griep,
S.H.Hinrichs,
and
P.Soultanas
(2007).
Domain swapping reveals that the C- and N-terminal domains of DnaG and DnaB, respectively, are functional homologues.
|
| |
Mol Microbiol,
63,
1629-1639.
|
 |
|
|
|
|
 |
S.Bailey,
W.K.Eliason,
and
T.A.Steitz
(2007).
Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase.
|
| |
Science,
318,
459-463.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Bailey,
W.K.Eliason,
and
T.A.Steitz
(2007).
The crystal structure of the Thermus aquaticus DnaB helicase monomer.
|
| |
Nucleic Acids Res,
35,
4728-4736.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.E.Corn,
and
J.M.Berger
(2006).
Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions.
|
| |
Nucleic Acids Res,
34,
4082-4088.
|
 |
|
|
|
|
 |
J.Thirlway,
and
P.Soultanas
(2006).
In the Bacillus stearothermophilus DnaB-DnaG complex, the activities of the two proteins are modulated by distinct but overlapping networks of residues.
|
| |
J Bacteriol,
188,
1534-1539.
|
 |
|
|
|
|
 |
X.C.Su,
P.M.Schaeffer,
K.V.Loscha,
P.H.Gan,
N.E.Dixon,
and
G.Otting
(2006).
Monomeric solution structure of the helicase-binding domain of Escherichia coli DnaG primase.
|
| |
FEBS J,
273,
4997-5009.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.E.Corn,
P.J.Pease,
G.L.Hura,
and
J.M.Berger
(2005).
Crosstalk between primase subunits can act to regulate primer synthesis in trans.
|
| |
Mol Cell,
20,
391-401.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Soultanas
(2005).
The bacterial helicase-primase interaction: a common structural/functional module.
|
| |
Structure,
13,
839-844.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

