 |
PDBsum entry 1ywo

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase, signaling protein
|
PDB id
|
|

|

|
1ywo
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase, signaling protein
|
 |
|
Title:
|
 |
Phospholipase cgamma1 sh3 in complex with a slp-76 motif
|
|
Structure:
|
 |
1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1. Chain: a. Synonym: phosphoinositide phospholipasE C, plc-gamma-1, phospholipasE C-gamma-1, plc-ii, plc-148. Engineered: yes. Mutation: yes. Lymphocyte cytosolic protein 2. Chain: p.
|
|
Source:
|
 |
Rattus norvegicus. Norway rat. Organism_taxid: 10116. Gene: plcg1. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008. Homo sapiens. Human. Organism_taxid: 9606
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
1.81Å
|
R-factor:
|
0.173
|
R-free:
|
0.221
|
|
|
Authors:
|
 |
L.Deng,C.A.Velikovsky,C.P.Swaminathan,S.Cho,R.A.Mariuzza
|
Key ref:

|
 |
L.Deng
et al.
(2005).
Structural basis for recognition of the T cell adaptor protein SLP-76 by the SH3 domain of phospholipase Cgamma1.
J Mol Biol,
352,
1.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
18-Feb-05
|
Release date:
|
16-Aug-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P10686
(PLCG1_RAT) -
1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 from Rattus norvegicus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1290 a.a.
55 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.11
- phosphoinositide phospholipase C.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
myo-Inositol Phosphate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate) + H2O = 1D-myo-inositol 1,4,5-trisphosphate + a 1,2-diacyl-sn-glycerol + H+
|
 |
 |
 |
 |
 |
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate)
|
+
|
H2O
|
=
|
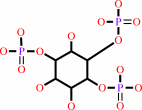 1D-myo-inositol 1,4,5-trisphosphate
1D-myo-inositol 1,4,5-trisphosphate
|
+
|
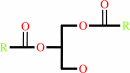 1,2-diacyl-sn-glycerol
1,2-diacyl-sn-glycerol
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
352:1
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for recognition of the T cell adaptor protein SLP-76 by the SH3 domain of phospholipase Cgamma1.
|
|
L.Deng,
C.A.Velikovsky,
C.P.Swaminathan,
S.Cho,
R.A.Mariuzza.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The enzyme phospholipase Cgamma1 (PLCgamma1) is essential for T cell signaling
and activation. Following T cell receptor ligation, PLCgamma1 interacts through
its SH2 and SH3 domains with the adaptors LAT and SLP-76, respectively, to form
a multiprotein signaling complex that leads to activation of PLCgamma1 by Syk
tyrosine kinases. To identify the binding site for PLCgamma1 in SLP-76, we used
isothermal titration calorimetry to measure affinities for the interaction of
PLCgamma1-SH3 with a set of overlapping peptides spanning the central
proline-rich region of SLP-76. PLCgamma1-SH3 bound with high specificity to the
SLP-76 motif 186PPVPPQRP193, which represents the minimal binding site. To
understand the basis for selective recognition, we determined the crystal
structures of PLCgamma1-SH3 in free form, and bound to a 10-mer peptide
containing this site, to resolutions of 1.60 A and 1.81 A, respectively. The
structures reveal that several key contacting residues of the SH3 shift toward
the SLP-76 peptide upon complex formation, optimizing the fit and strengthening
hydrophobic interactions. Selectivity results mainly from strict shape
complementarity between protein and peptide, rather than sequence-specific
hydrogen bonding. In addition, Pro193 of SLP-76 assists in positioning Arg192
into the compass pocket of PLCgamma1-SH3, which coordinates the compass residue
through an unusual aspartate. The PLCgamma1-SH3/SLP-76 structure provides
insights into ligand binding by SH3 domains related to PLCgamma1-SH3, as well as
into recognition by PLCgamma1 of signaling partners other than SLP-76.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Comparison of PLCg1-SH3 in free and SLP-76-bound
forms. (a) Superposition of the polypeptide chain of unbound
PLCg1-SH3 (gold) onto that of PLCg1-SH3 in the PLCg1-SH3/SLP-76
complex (cyan). The SLP-76 peptide is drawn in ball-and-stick
representation. The arrow points to the region where the two
structures deviate most. (b) Close-up view of the rearrangement
of the n-Src loop of PLCg1-SH3 following peptide binding.
Glycine residues are represented by their carbonyl oxygen atoms.
(c) Adjustments in the PLCg1-SH3 binding site upon ligation of
SLP-76 peptide. Free PLCg1-SH3 was superposed on SLP-76-bound
PLCg1-SH3. The view is looking down on the binding groove. The
SLP-76 peptide and the rest of the SH3 structure are omitted for
clarity. Only residues contacting the ligand are shown. Glycine
residues are represented by their carbonyl oxygen atoms. Dual
conformations were observed for the side-chains Gln805 and
Arg806 in the PLCg1-SH3/SLP-76 complex.
|
 |
Figure 4.
Figure 4. (a) Structure-based sequence alignment of the SH3
domains of PLCg1, PLCg2 and Lck. Secondary structure elements
are denoted by violet arrows (b-strands) and a pink cylinder
(3[10] helix). These, and the loop regions (brown lines), are
labeled according to the nomenclature for Src-SH3.22 Residues of
PLCg1-SH3 making van der Waals contacts with SLP-76 are shaded
yellow; residues interacting with SLP-76 through hydrogen bonds
are shaded cyan. Asp808, which forms a salt-bridge with the
bound peptide, is shaded red. (b) Superposition of PLCg1-SH3 in
bound form (cyan) onto free Lck-SH3 (green; PDB accession code
1LCK).28 The view is the same as that in Figure 3(c). Residues
of Lck-SH3 that potentially contact bound peptide, based on
sequence alignment with PLCg1-SH3 (a), are shown. (c)
Interactions at the SH3 compass pocket. The PLCg1/SLP-76 complex
(cyan) was superposed onto the Src-SH3/App12 complex (PDB
accession code 1QWE)30 and free Lck-SH3. Only interactions with
the compass residue of the bound peptide (Arg192 of SLP-76 and
Arg9 of App12) are shown. Salt-bridges are drawn as broken lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2005,
352,
1-0)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Buch,
D.Fishelovitch,
N.London,
B.Raveh,
H.J.Wolfson,
and
R.Nussinov
(2010).
Allosteric regulation of glycogen synthase kinase 3β: a theoretical study.
|
| |
Biochemistry,
49,
10890-10901.
|
 |
|
|
|
|
 |
A.Ababou,
and
J.E.Ladbury
(2007).
Survey of the year 2005: literature on applications of isothermal titration calorimetry.
|
| |
J Mol Recognit,
20,
4.
|
 |
|
|
|
|
 |
Q.Qi,
and
A.August
(2007).
Keeping the (kinase) party going: SLP-76 and ITK dance to the beat.
|
| |
Sci STKE,
2007,
pe39.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

