 |
PDBsum entry 1xhx
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Insights into strand displacement and processivity from the crystal structure of the protein-Primed DNA polymerase of bacteriophage phi29.
|
 |
|
Authors
|
 |
S.Kamtekar,
A.J.Berman,
J.Wang,
J.M.Lázaro,
M.De vega,
L.Blanco,
M.Salas,
T.A.Steitz.
|
 |
|
Ref.
|
 |
Mol Cell, 2004,
16,
609-618.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
The DNA polymerase from phage phi29 is a B family polymerase that initiates
replication using a protein as a primer, attaching the first nucleotide of the
phage genome to the hydroxyl of a specific serine of the priming protein. The
crystal structure of phi29 DNA polymerase determined at 2.2 A resolution
provides explanations for its extraordinary processivity and strand displacement
activities. Homology modeling suggests that downstream template DNA passes
through a tunnel prior to entering the polymerase active site. This tunnel is
too small to accommodate double-stranded DNA and requires the separation of
template and nontemplate strands. Members of the B family of DNA polymerases
that use protein primers contain two sequence insertions: one forms a domain not
previously observed in polymerases, while the second resembles the specificity
loop of T7 RNA polymerase. The high processivity of phi29 DNA polymerase may be
explained by its topological encirclement of both the downstream template and
the upstream duplex DNA.
|
 |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Ribbon Representation of the Domain Organization
of φ29 DNA PolymeraseThe exonuclease domain is shown in red,
the palm in pink, TPR1 in gold, the fingers in blue, TPR2 in
cyan, and the thumb in green. D249 and D458, which provide the
catalytic carboxylates of the polymerase active site, are shown
using space-filling spheres.
|
 |
Figure 4.
Figure 4. Structures of TPR1 and TPR2, Domains that Are
Specific to Protein-Primed DNA Polymerases(A) TPR1 forms a
compact domain. This region is an insertion between the palm and
the fingers subdomains. The motif, identified on the basis of
sequence analysis (residues 302–358, gold), can be extended to
include residues 261–301 as well (brown), thereby forming a
subdomain with no homology to the palm subdomains of other B
family polymerases.(B) Structural analogy between TPR2 (cyan)
and the specificity loop (gold) of T7 RNA polymerase. The
fragments of both palms used for superposition are colored in
pink (φ29 DNA polymerase) and gray (T7 RNA polymerase). The
atoms of the residues containing the catalytic carboxylates are
shown as space-filling spheres.
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from Cell Press:
Mol Cell
(2004,
16,
609-618)
copyright 2004.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
Correction of X-Ray intensities from single crystals containing lattice-Translocation defects.
|
 |
|
Authors
|
 |
J.Wang,
S.Kamtekar,
A.J.Berman,
T.A.Steitz.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2005,
61,
67-74.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
|
Note In the PDB file this reference is
annotated as "TO BE PUBLISHED".
The citation details given above were identified by an automated
search of PubMed on title and author
names, giving a
percentage match of
88%.
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 6.
Figure 6 Reciprocal-space lattice in the presence of the
lattice-translocation defect of Fig. 3-with 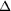 z
= 1/2. When the defect occurs with low frequency ( z
= 1/2. When the defect occurs with low frequency (  <
5%), the streaky features are limited to slightly elliptically
shaped lattice points. Such a limited defect can be partially
corrected during intensity integration using an elliptical
spot-shape option when the crystal is rotated along the a* axis
during data collection, but it cannot when the axis of the
crystal rotation is perpendicular to a*. In the latter case,
streaky features would appear only in one orientation and many
extra Bragg reflections would not be predictable with a standard
definition of the mosaicity in a second orientation that is
90° away from the first orientation. <
5%), the streaky features are limited to slightly elliptically
shaped lattice points. Such a limited defect can be partially
corrected during intensity integration using an elliptical
spot-shape option when the crystal is rotated along the a* axis
during data collection, but it cannot when the axis of the
crystal rotation is perpendicular to a*. In the latter case,
streaky features would appear only in one orientation and many
extra Bragg reflections would not be predictable with a standard
definition of the mosaicity in a second orientation that is
90° away from the first orientation.
|
 |
Figure 7.
Figure 7 An experimental electron-density map of 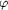 29
DNA polymerase at 2.5 Å using phases initially derived
from Hg derivatives after their X-ray intensities were
corrected, followed by density modification and twofold
non-crystallographic symmetry averaging. This map is contoured
at 1.0 29
DNA polymerase at 2.5 Å using phases initially derived
from Hg derivatives after their X-ray intensities were
corrected, followed by density modification and twofold
non-crystallographic symmetry averaging. This map is contoured
at 1.0  and
superimposed with the finally refined model. and
superimposed with the finally refined model.
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from the IUCr
|
 |
|

|
|
|
 |

