 |
PDBsum entry 1uot

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Regulator of complement pathway
|
PDB id
|
|

|

|
1uot
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Mapping cd55 function. The structure of two pathogen-Binding domains at 1.7 a.
|
 |
|
Authors
|
 |
P.Williams,
Y.Chaudhry,
I.G.Goodfellow,
J.Billington,
R.Powell,
O.B.Spiller,
D.J.Evans,
S.Lea.
|
 |
|
Ref.
|
 |
J Biol Chem, 2003,
278,
10691-10696.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
Decay-accelerating factor (CD55), a regulator of the alternative and classical
pathways of complement activation, is expressed on all serum-exposed cells. It
is used by pathogens, including many enteroviruses and uropathogenic Escherichia
coli, as a receptor prior to infection. We describe the x-ray structure of a
pathogen-binding fragment of human CD55 at 1.7 A resolution containing two of
the three domains required for regulation of human complement. We have used
mutagenesis to map biological functions onto the molecule; decay-accelerating
activity maps to a single face of the molecule, whereas bacterial and viral
pathogens recognize a variety of different sites on CD55.
|
 |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Fold and arrangement of SCR domains in CD55[34].
This figure was drawn using AESOP (M. E. M. Noble, unpublished
program). A, secondary structure and location of disulfide bonds
in CD55[34]. Strands are labeled according to the convention
defined by Norman et al. (35). Note that there is no strand 1 as
the hydrogen-bonding pattern of these residues does not meet the
strict criteria for definition of a  -strand. B,
variation in orientation between SCR domains 3 and 4 in the five
independent copies of CD55[34] found in the different crystal
forms (Table I). C, model for topology of CD55 in the membrane
based on our structure of CD55[34]. -strand. B,
variation in orientation between SCR domains 3 and 4 in the five
independent copies of CD55[34] found in the different crystal
forms (Table I). C, model for topology of CD55 in the membrane
based on our structure of CD55[34].
|
 |
Figure 3.
Fig. 3. Mapping of functional data onto the structure of
CD55[34]. This figure was drawn using AESOP (M. E. M. Noble,
unpublished program). All plates show two views of the surface
of CD55, the first view corresponding to the orientation as
shown in Fig. 1 and termed Front View. The Back View corresponds
to a rotation of 180 degrees about the long axis of the
molecule. A, orange indicates the sites mutagenized in this
study, and yellow indicates the sites of sequence difference
between AGM and human CD55. B, AP indicates the mutation shown
to effect alternative pathway decay acceleration, and CP
indicates those sites shown to affect classical pathway decay
acceleration; both those sites have been shown to affect both
pathways. Dark green indicates those sites shown previously to
effect both classical and alternative pathway decay acceleration
(22). C, sites marked with a virus name have been shown to
affect binding of that virus. Sites marked in pale pink are the
sites of difference between AGM and human CD55 that are known to
abolish EV11 binding and significantly reduce EV12 binding. D,
the locations of two of the Cromer variants of human CD55 are
shown. Different E. coli strains are sensitive to changes at one
or other of these positions (28, 30, 32).
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
10691-10696)
copyright 2003.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
Crystallization and preliminary X-Ray diffraction analysis of a biologically active fragment of cd55.
|
 |
|
Authors
|
 |
S.Lea,
R.Powell,
D.Evans.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 1999,
55,
1198-1200.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |
Figure 1.
Figure 1 Crystals of CD55 domains 3 and 4. (a) Orthorhombic
crystal form; crystal is 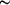 0.5
mm in the longest dimension. (b) Monoclinic crystal form;
crystal on the right is 0.5
mm in the longest dimension. (b) Monoclinic crystal form;
crystal on the right is 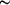 0.3
mm in the longest dimension. 0.3
mm in the longest dimension.
|
 |
|
 |
 |
|
The above figure is
reproduced from the cited reference
with permission from the IUCr
|
 |
|
Secondary reference #2
|
 |
|
Title
|
 |
Determination of the affinity and kinetic constants for the interaction between the human virus echovirus 11 and its cellular receptor, Cd55.
|
 |
|
Authors
|
 |
S.M.Lea,
R.M.Powell,
T.Mckee,
D.J.Evans,
D.Brown,
D.I.Stuart,
P.A.Van der merwe.
|
 |
|
Ref.
|
 |
J Biol Chem, 1998,
273,
30443-30447.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |
Figure 1.
Fig. 1. Measuring the affinity and kinetics of DAF[234]
binding to EV11 by surface plasmon resonance. A, DAF[234] (12
µM) was injected (bar) for 22.5 s over sensor surfaces
with a high level of (3020 RU), a low level of (1370 RU), or no
(control) EV11 immobilized. An overlay plot of all three
sensorgrams is shown, normalized to the same preinjection level.
Inset, same sensorgrams after subtraction of the control
sensorgram. In B, DAF[234] was injected (bar) at the indicated
concentration for 22.5 s over sensor surfaces with EV11 (3020
RU) or no protein immobilized. An overlay plot is shown of all
five sensorgrams after subtraction of their respective control
sensorgrams. Curves, a fit of the simple Langmuir binding model
(A + B  AB) to all
sensorgrams simultaneously gives a k[on] of 140,000 M AB) to all
sensorgrams simultaneously gives a k[on] of 140,000 M  1·s 1·s
 1 and a
k[off] of 0.3 s 1 and a
k[off] of 0.3 s  1. The
curve fitting was performed by numerical integration using the
global analysis option (i.e. simultaneous fitting of the
association and dissociation phases) of BIAevaluation 3.0
(BIAcore AB). C, plot of the equilibrium response obtained in A
during injection of different DAF[234] concentrations. Nonlinear
curve fitting of the Langmuir binding model to these data (bar)
gives a K[D] of 2.8 µM. Inset, Scatchard plot of the same
data. A linear fit of these data (bar) also gives a K[D] of 2.8
µM. 1. The
curve fitting was performed by numerical integration using the
global analysis option (i.e. simultaneous fitting of the
association and dissociation phases) of BIAevaluation 3.0
(BIAcore AB). C, plot of the equilibrium response obtained in A
during injection of different DAF[234] concentrations. Nonlinear
curve fitting of the Langmuir binding model to these data (bar)
gives a K[D] of 2.8 µM. Inset, Scatchard plot of the same
data. A linear fit of these data (bar) also gives a K[D] of 2.8
µM.
|
 |
|
 |
 |
|
The above figure is
reproduced from the cited reference
with permission from the ASBMB
|
 |
|

|
|
|
 |

