 |
PDBsum entry 1t8o

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/hydrolase inhibitor
|
PDB id
|
|

|

|
1t8o
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Crystal structures of five bovine chymotrypsin complexes with p1 bpti variants.
|
 |
|
Authors
|
 |
H.Czapinska,
R.Helland,
A.O.Smalås,
J.Otlewski.
|
 |
|
Ref.
|
 |
J Mol Biol, 2004,
344,
1005-1020.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
The bovine chymotrypsin-bovine pancreatic trypsin inhibitor (BPTI) interaction
belongs to extensively studied models of protein-protein recognition. The
accommodation of the inhibitor P1 residue in the S1 binding site of the enzyme
forms the hot spot of this interaction. Mutations introduced at the P1 position
of BPTI result in a more than five orders of magnitude difference of the
association constant values with the protease. To elucidate the structural
aspects of the discrimination between different P1 residues, crystal structures
of five bovine chymotrypsin-P1 BPTI variant complexes have been determined at pH
7.8 to a resolution below 2 A. The set includes polar (Thr), ionizable (Glu,
His), medium-sized aliphatic (Met) and large aromatic (Trp) P1 residues and
complements our earlier studies of the interaction of different P1 side-chains
with the S1 pocket of chymotrypsin. The structures have been compared to the
complexes of proteases with similar and dissimilar P1 preferences, including
Streptomyces griseus proteases B and E, human neutrophil elastase, crab
collagenase, bovine trypsin and human thrombin. The S1 sites of these enzymes
share a common general shape of significant rigidity. Large and branched P1
residues adapt in their complexes similar conformations regardless of the
polarity and size differences between their S1 pockets. Conversely, long and
flexible residues such as P1 Met are present in the disordered form and display
a conformational diversity despite similar inhibitory properties with respect to
most enzymes studied. Thus, the S1 specificity profiles of the serine proteases
appear to result from the precise complementarity of the P1-S1 interface and
minor conformational adjustments occurring upon the inhibitor binding.
|
 |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The electron density for the P1 residue of BPTI
and residues Ser189, Ser190 and Met192 of chymotrypsin together
with the hydrogen bonding pattern of the P1 side-chain in the
structures of the following complexes: (a) Chtp-K15E BPTI; (b)
Chtp-K15M BPTI; (c) Chtp-K15H BPTI; (d) Chtp-K15T BPTI; (e)
Chtp-K15W BPTI. The 2F[o] -F[c] maps were contoured at 1.5s.
Selected side-chains are indicated in red, the binding loop of
the inhibitor in orange and the S1 binding pocket of the enzyme
in gray (only the main-chain of the S1 pocket together with
Cys191 and Cys220 side-chains is presented for clarity). This
and the following Figures were produced with the program
XtalView.47
|
 |
Figure 6.
Figure 6. The superposition of the S1 pockets in P1 Trp
BPTI-bovine chymotrypsin complex (red/orange); P1 Trp
BPTI-bovine trypsin complex (PDB 3BTW, blue); and the complex of
thrombin with the P1 Trp possessing peptidyl inhibitor (PDB
1AD8, green).
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2004,
344,
1005-1020)
copyright 2004.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
Structural consequences of accommodation of four non-Cognate amino acid residues in the s1 pocket of bovine trypsin and chymotrypsin.
|
 |
|
Authors
|
 |
R.Helland,
H.Czapinska,
I.Leiros,
M.Olufsen,
J.Otlewski,
A.O.Smalås.
|
 |
|
Ref.
|
 |
J Mol Biol, 2003,
333,
845-861.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. Electron density of the P1 residue, Met192 and
solvent molecules in the chymotrypsin-BPTI complexes: (a) P1
Gly, (b) P1 Val, (c) P1 Leu and (d) P1 Phe: 2F[o] -F[c] density
is colored blue at 1s, and F[o] -F[c] density is colored green
at 3s.
|
 |
Figure 6.
Figure 6. P1 Gly BPTI-binding epitope (residues 11-19 and
38-40) on the electrostatic surface of trypsin (left) and
chymotrypsin (right). The Figure was made using ICM (MolSoft
LLC, La Jolla).
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from Elsevier
|
 |
|
Secondary reference #2
|
 |
|
Title
|
 |
Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of alzheimer'S amyloid beta-Protein precursor (appi) and basic pancreatic trypsin inhibitor (bpti): engineering of inhibitors with altered specificities.
|
 |
|
Authors
|
 |
A.J.Scheidig,
T.R.Hynes,
L.A.Pelletier,
J.A.Wells,
A.A.Kossiakoff.
|
 |
|
Ref.
|
 |
Protein Sci, 1997,
6,
1806-1824.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
|
Secondary reference #3
|
 |
|
Title
|
 |
Crystal structure of the bovine alpha-Chymotrypsin:kunitz inhibitor complex. An example of multiple protein:protein recognition sites.
|
 |
|
Authors
|
 |
C.Capasso,
M.Rizzi,
E.Menegatti,
P.Ascenzi,
M.Bolognesi.
|
 |
|
Ref.
|
 |
J Mol Recognit, 1997,
10,
26-35.
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
|
Secondary reference #4
|
 |
|
Title
|
 |
Ultrahigh-Resolution structure of a bpti mutant.
|
 |
|
Authors
|
 |
A.Addlagatta,
S.Krzywda,
H.Czapinska,
J.Otlewski,
M.Jaskolski.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2001,
57,
649-663.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2 Molecular interactions of Tyr35. The  electrons
of the aromatic ring accept two N-H hydrogen bonds from the
main-chain amide of Gly37 and from the side-chain amide of
Asn44. The OH group of Tyr35 is also involved in an
intermolecular hydrogen bond to a sulfate anion. (a) Electron
density of the interacting residues: 3F[o] - 2F[c] map contoured
at 0.65 e Å-3 (1.7 electrons
of the aromatic ring accept two N-H hydrogen bonds from the
main-chain amide of Gly37 and from the side-chain amide of
Asn44. The OH group of Tyr35 is also involved in an
intermolecular hydrogen bond to a sulfate anion. (a) Electron
density of the interacting residues: 3F[o] - 2F[c] map contoured
at 0.65 e Å-3 (1.7  ,
blue) and H-omit difference map contoured at 0.10 e Å-3 (2.5 ,
blue) and H-omit difference map contoured at 0.10 e Å-3 (2.5
 ,
green); (b) the same fragment shown with anisotropic
displacement ellipsoids drawn at 20% probability. Hydrogen-bond
distances (Å) are from the H atoms to the centre of the aromatic
ring. ,
green); (b) the same fragment shown with anisotropic
displacement ellipsoids drawn at 20% probability. Hydrogen-bond
distances (Å) are from the H atoms to the centre of the aromatic
ring.
|
 |
Figure 4.
Figure 4 C=O  C=O
dipole interactions between the side-chain and main-chain
carbonyl groups of the three Asn residues. Those residues are
among the best regions of the structure, as illustrated by the
3F[o] - 2F[c] electron density. Contour level: 2.01 e Å-3 (5.3 C=O
dipole interactions between the side-chain and main-chain
carbonyl groups of the three Asn residues. Those residues are
among the best regions of the structure, as illustrated by the
3F[o] - 2F[c] electron density. Contour level: 2.01 e Å-3 (5.3
 )
for Asn24 and 2.12 e Å-3 (5.7 )
for Asn24 and 2.12 e Å-3 (5.7  )
for Asn43-Asn44. The atom-atom distances in these interactions
are given in Å and are indicated by dotted lines. The
conformation of the main chain is described by the )
for Asn43-Asn44. The atom-atom distances in these interactions
are given in Å and are indicated by dotted lines. The
conformation of the main chain is described by the 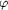 / /
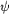 values
and that of the side chains by the values
and that of the side chains by the 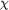 [1]
torsion angles. [1]
torsion angles.
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from the IUCr
|
 |
|
Secondary reference #5
|
 |
|
Title
|
 |
Crystallographic refinement of the structure of bovine pancreatic trypsin inhibitor at 1.5 a resolution
|
 |
|
Authors
|
 |
J.Deisenhofer,
W.Steigemann.
|
 |
|
Ref.
|
 |
acta crystallogr , sect b, 1975,
31,
238.
|
 |
 |
|
Secondary reference #6
|
 |
|
Title
|
 |
Three-Dimensional structure of tosyl-Alpha-Chymotrypsin.
|
 |
|
Authors
|
 |
B.W.Matthews,
P.B.Sigler,
R.Henderson,
D.M.Blow.
|
 |
|
Ref.
|
 |
Nature, 1967,
214,
652-656.
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
|

|
|
|
 |

