 |
PDBsum entry 1t0t

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics, unknown function
|
PDB id
|
|

|

|
1t0t
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Structural genomics, unknown function
|
 |
|
Title:
|
 |
Crystallographic structure of a putative chlorite dismutase
|
|
Structure:
|
 |
Apc35880. Chain: v, w, x, y, z. Engineered: yes
|
|
Source:
|
 |
Geobacillus stearothermophilus. Organism_taxid: 1422
|
|
Biol. unit:
|
 |
Pentamer (from
)
|
|
Resolution:
|
 |
|
1.75Å
|
R-factor:
|
0.158
|
R-free:
|
0.194
|
|
|
Authors:
|
 |
M.Gilski,D.Borek,Y.Chen,F.Collart,A.Joachimiak,Z.Otwinowski,Midwest Center For Structural Genomics (Mcsg)
|
|
Key ref:
|
 |
M.Gilski
et al.
Crystal structure of apc35880 protein from bacillus stearothermophilus.
To be published,
.
|
 |
|
Date:
|
 |
|
12-Apr-04
|
Release date:
|
24-Aug-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5KUD5
(Y3416_GEOKA) -
Coproheme decarboxylase from Geobacillus kaustophilus (strain HTA426)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
248 a.a.
243 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.98.5
- hydrogen peroxide-dependent heme synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Fe-coproporphyrin III + 2 H2O2 + 2 H+ = heme b + 2 CO2 + 4 H2O
|
 |
 |
 |
 |
 |
Fe-coproporphyrin III
|
+
|
 2
×
H2O2
2
×
H2O2
|
+
|
2
×
H(+)
|
=
|
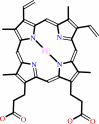 heme b
heme b
|
+
|
 2
×
CO2
2
×
CO2
|
+
|
4
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

