 |
PDBsum entry 1r0l

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1r0l
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
1-deoxy-d-xylulose 5-phosphate reductoisomerase from zymomonas mobilis in complex with NADPH
|
|
Structure:
|
 |
1-deoxy-d-xylulose 5-phosphate reductoisomerase. Chain: a, b, c, d. Synonym: dxp reductoisomerase, 1-deoxyxylulose-5-phosphate reductoisomerase. Engineered: yes
|
|
Source:
|
 |
Zymomonas mobilis. Organism_taxid: 542. Gene: dxr. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.249
|
R-free:
|
0.269
|
|
|
Authors:
|
 |
S.Ricagno,S.Grolle,S.Bringer-Meyer,H.Sahm,Y.Lindqvist,G.Schneider
|
|
Key ref:
|
 |
S.Ricagno
et al.
(2004).
Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim Biophys Acta,
1698,
37-44.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
22-Sep-03
|
Release date:
|
13-Jul-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9X5F2
(DXR_ZYMMO) -
1-deoxy-D-xylulose 5-phosphate reductoisomerase from Zymomonas mobilis subsp. mobilis (strain ATCC 31821 / ZM4 / CP4)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
388 a.a.
378 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.267
- 1-deoxy-D-xylulose-5-phosphate reductoisomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2-C-methyl-D-erythritol 4-phosphate + NADP+ = 1-deoxy-D-xylulose 5-phosphate + NADPH + H+
|
 |
 |
 |
 |
 |
 2-C-methyl-D-erythritol 4-phosphate
2-C-methyl-D-erythritol 4-phosphate
|
+
|
NADP(+)
Bound ligand (Het Group name = )
matches with 56.25% similarity
|
=
|
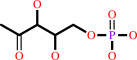 1-deoxy-D-xylulose 5-phosphate
1-deoxy-D-xylulose 5-phosphate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+) or cobalt cation or Mn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochim Biophys Acta
1698:37-44
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
|
|
S.Ricagno,
S.Grolle,
S.Bringer-Meyer,
H.Sahm,
Y.Lindqvist,
G.Schneider.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
1-Deoxy-d-xylulose-5-phosphate reductoisomerase (DXR) is the second enzyme in
the non-mevalonate pathway of isoprenoid biosynthesis. The structure of the
apo-form of this enzyme from Zymomonas mobilis has been solved and refined to
1.9-A resolution, and that of a binary complex with the co-substrate NADPH to
2.7-A resolution. The subunit of DXR consists of three domains. Residues 1-150
form the NADPH binding domain, which is a variant of the typical
dinucleotide-binding fold. The second domain comprises a four-stranded mixed
beta-sheet, with three helices flanking the sheet. Most of the putative active
site residues are located on this domain. The C-terminal domain (residues
300-386) folds into a four-helix bundle. In solution and in the crystal, the
enzyme forms a homo-dimer. The interface between the two monomers is formed
predominantly by extension of the sheet in the second domain. The adenosine
phosphate moiety of NADPH binds to the nucleotide-binding fold in the canonical
way. The adenine ring interacts with the loop after beta1 and with the loops
between alpha2 and beta2 and alpha5 and beta5. The nicotinamide ring is
disordered in crystals of this binary complex. Comparisons to Escherichia coli
DXR show that the two enzymes are very similar in structure, and that the active
site architecture is highly conserved. However, there are differences in the
recognition of the adenine ring of NADPH in the two enzymes.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.L.Williams,
and
J.Andrew McCammon
(2009).
Conformational Dynamics of the Flexible Catalytic Loop in Mycobacterium tuberculosis 1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase.
|
| |
Chem Biol Drug Des,
73,
26-38.
|
 |
|
|
|
|
 |
L.M.Henriksson,
T.Unge,
J.Carlsson,
J.Aqvist,
S.L.Mowbray,
and
T.A.Jones
(2007).
Structures of Mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate reductoisomerase provide new insights into catalysis.
|
| |
J Biol Chem,
282,
19905-19916.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Singh,
M.A.Avery,
and
C.R.McCurdy
(2007).
Toward Mycobacterium tuberculosis DXR inhibitor design: homology modeling and molecular dynamics simulations.
|
| |
J Comput Aided Mol Des,
21,
511-522.
|
 |
|
|
|
|
 |
S.Yajima,
K.Hara,
D.Iino,
Y.Sasaki,
T.Kuzuyama,
K.Ohsawa,
and
H.Seto
(2007).
Structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in a quaternary complex with a magnesium ion, NADPH and the antimalarial drug fosmidomycin.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
466-470.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.N.Hunter
(2007).
The non-mevalonate pathway of isoprenoid precursor biosynthesis.
|
| |
J Biol Chem,
282,
21573-21577.
|
 |
|
|
|
|
 |
L.M.Henriksson,
C.Björkelid,
S.L.Mowbray,
and
T.Unge
(2006).
The 1.9 A resolution structure of Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate reductoisomerase, a potential drug target.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
807-813.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Wiesner,
and
F.Seeber
(2005).
The plastid-derived organelle of protozoan human parasites as a target of established and emerging drugs.
|
| |
Expert Opin Ther Targets,
9,
23-44.
|
 |
|
|
|
|
 |
L.Mercklé,
A.de Andrés-Gómez,
B.Dick,
R.J.Cox,
and
C.R.Godfrey
(2005).
A fragment-based approach to understanding inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase.
|
| |
Chembiochem,
6,
1866-1874.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

